|
Symbol Name ID |
Ush1g
USH1 protein network component sans MGI:2450757 |
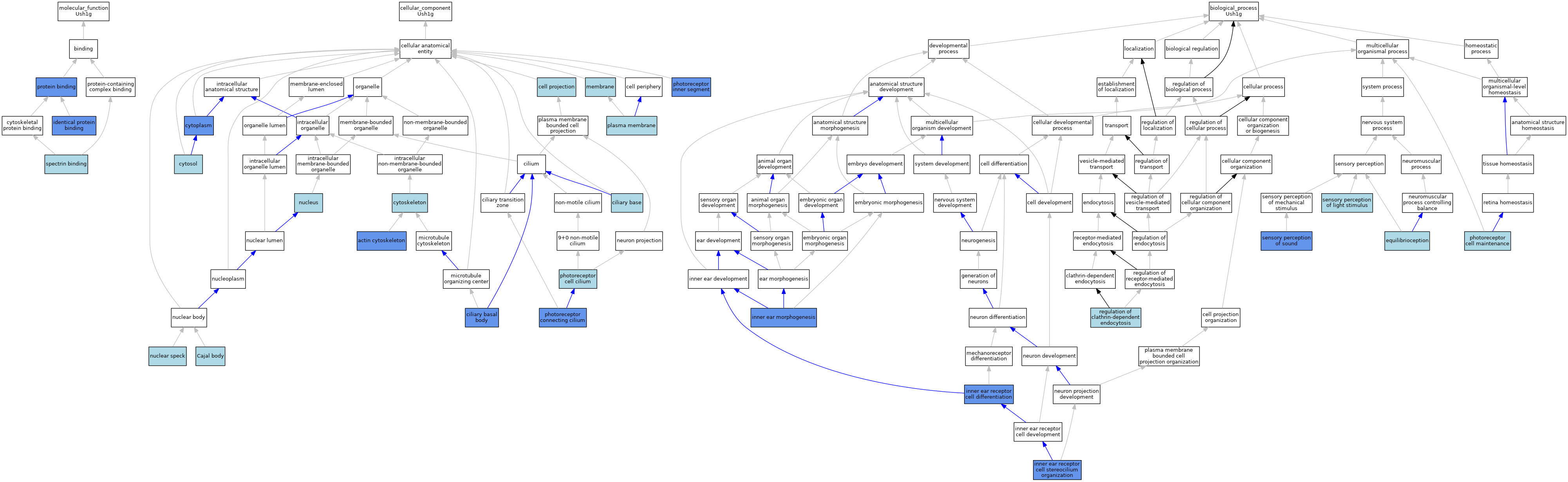
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:95929 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:95929 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:95929 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:132040 | |||||||||
| Molecular Function | GO:0030507 | spectrin binding | ISO | J:199579 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | IDA | J:82023 | |||||||||
| Cellular Component | GO:0015030 | Cajal body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0036064 | ciliary basal body | IDA | J:132040 | |||||||||
| Cellular Component | GO:0097546 | ciliary base | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:95929 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9659536 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9663360 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISS | J:82639 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0097733 | photoreceptor cell cilium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032391 | photoreceptor connecting cilium | IDA | J:132040 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IDA | J:132040 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0050957 | equilibrioception | ISO | J:164563 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IMP | J:82022 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IMP | J:1174 | |||||||||
| Biological Process | GO:0060113 | inner ear receptor cell differentiation | IMP | J:1174 | |||||||||
| Biological Process | GO:0060113 | inner ear receptor cell differentiation | IMP | J:82022 | |||||||||
| Biological Process | GO:0060113 | inner ear receptor cell differentiation | IMP | J:3262 | |||||||||
| Biological Process | GO:0060113 | inner ear receptor cell differentiation | IMP | J:2196 | |||||||||
| Biological Process | GO:0060122 | inner ear receptor cell stereocilium organization | IMP | J:135991 | |||||||||
| Biological Process | GO:0045494 | photoreceptor cell maintenance | ISO | J:164563 | |||||||||
| Biological Process | GO:2000369 | regulation of clathrin-dependent endocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0050953 | sensory perception of light stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | ISO | J:164563 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | IMP | J:82022 | |||||||||
| Biological Process | GO:0007605 | sensory perception of sound | IMP | J:1174 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||