|
Symbol Name ID |
Ccdc40
coiled-coil domain containing 40 MGI:2443893 |
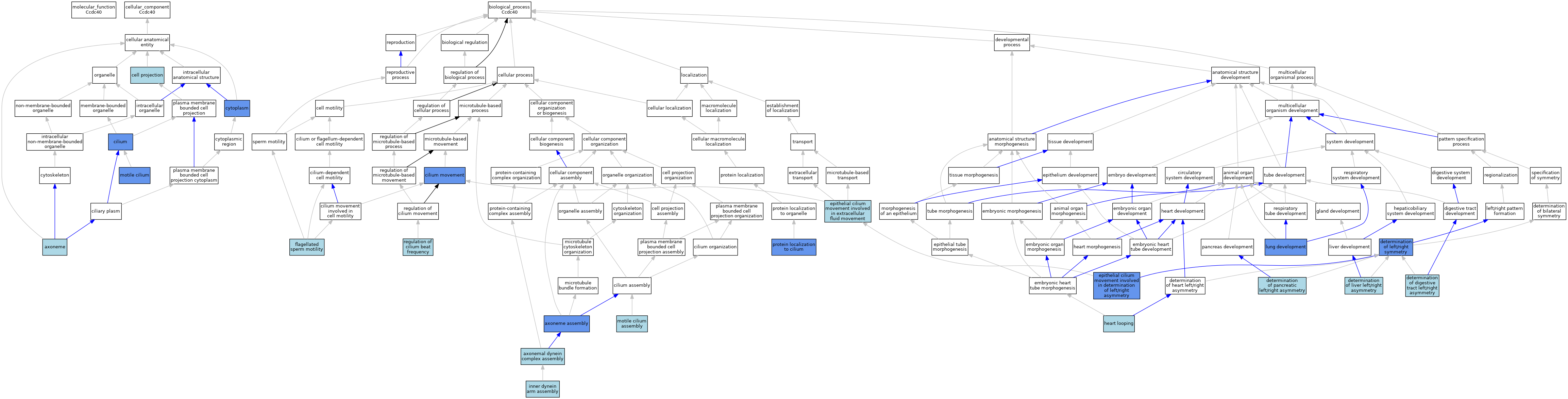
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular Component | GO:0005930 | axoneme | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005929 | cilium | IDA | J:168691 | |||||||||
| Cellular Component | GO:0005929 | cilium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005929 | cilium | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:168691 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031514 | motile cilium | IDA | J:230755 | |||||||||
| Biological Process | GO:0070286 | axonemal dynein complex assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0035082 | axoneme assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0035082 | axoneme assembly | IMP | J:230755 | |||||||||
| Biological Process | GO:0003341 | cilium movement | IMP | J:168691 | |||||||||
| Biological Process | GO:0003341 | cilium movement | ISO | J:164563 | |||||||||
| Biological Process | GO:0071907 | determination of digestive tract left/right asymmetry | ISO | J:164563 | |||||||||
| Biological Process | GO:0007368 | determination of left/right symmetry | IGI | J:245906 | |||||||||
| Biological Process | GO:0071910 | determination of liver left/right asymmetry | ISO | J:164563 | |||||||||
| Biological Process | GO:0035469 | determination of pancreatic left/right asymmetry | ISO | J:164563 | |||||||||
| Biological Process | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | ISO | J:164563 | |||||||||
| Biological Process | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | IMP | J:168691 | |||||||||
| Biological Process | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | IBA | J:265628 | |||||||||
| Biological Process | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | ISO | J:164563 | |||||||||
| Biological Process | GO:0030317 | flagellated sperm motility | ISO | J:164563 | |||||||||
| Biological Process | GO:0001947 | heart looping | ISO | J:164563 | |||||||||
| Biological Process | GO:0001947 | heart looping | IBA | J:265628 | |||||||||
| Biological Process | GO:0036159 | inner dynein arm assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0030324 | lung development | IMP | J:168691 | |||||||||
| Biological Process | GO:0030324 | lung development | ISO | J:164563 | |||||||||
| Biological Process | GO:0044458 | motile cilium assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0061512 | protein localization to cilium | IMP | J:230755 | |||||||||
| Biological Process | GO:0003356 | regulation of cilium beat frequency | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||