|
Symbol Name ID |
Osbpl8
oxysterol binding protein-like 8 MGI:2443807 |
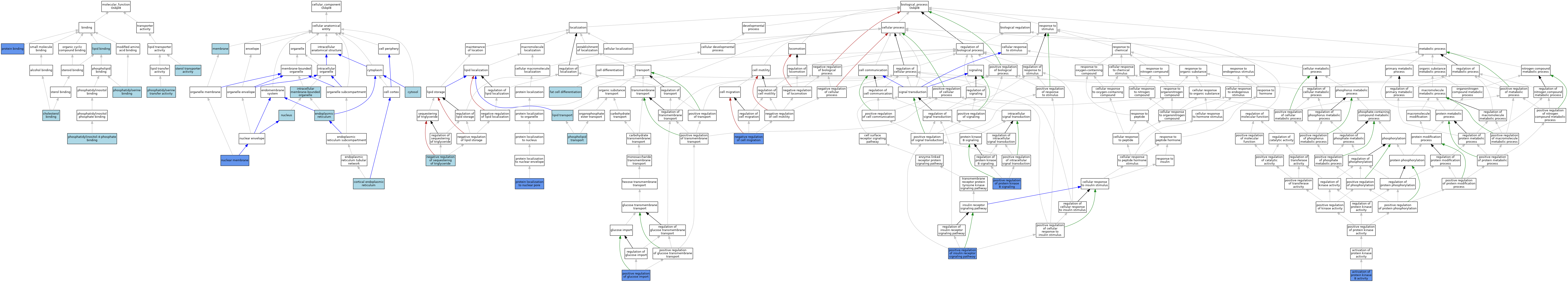
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0015485 | cholesterol binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015485 | cholesterol binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0070273 | phosphatidylinositol-4-phosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001786 | phosphatidylserine binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140343 | phosphatidylserine transfer activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:176266 | |||||||||
| Molecular Function | GO:0015248 | sterol transporter activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032541 | cortical endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | IMP | J:187070 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Biological Process | GO:0032148 | activation of protein kinase B activity | IMP | J:222138 | |||||||||
| Biological Process | GO:0045444 | fat cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006869 | lipid transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0030336 | negative regulation of cell migration | IMP | J:176266 | |||||||||
| Biological Process | GO:0010891 | negative regulation of sequestering of triglyceride | ISO | J:164563 | |||||||||
| Biological Process | GO:0015914 | phospholipid transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0046326 | positive regulation of glucose import | IMP | J:222138 | |||||||||
| Biological Process | GO:0046628 | positive regulation of insulin receptor signaling pathway | IDA | J:222138 | |||||||||
| Biological Process | GO:0046628 | positive regulation of insulin receptor signaling pathway | IMP | J:176266 | |||||||||
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | IMP | J:222138 | |||||||||
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | IMP | J:176266 | |||||||||
| Biological Process | GO:0090204 | protein localization to nuclear pore | IMP | J:176266 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||