|
Symbol Name ID |
Nceh1
neutral cholesterol ester hydrolase 1 MGI:2443191 |
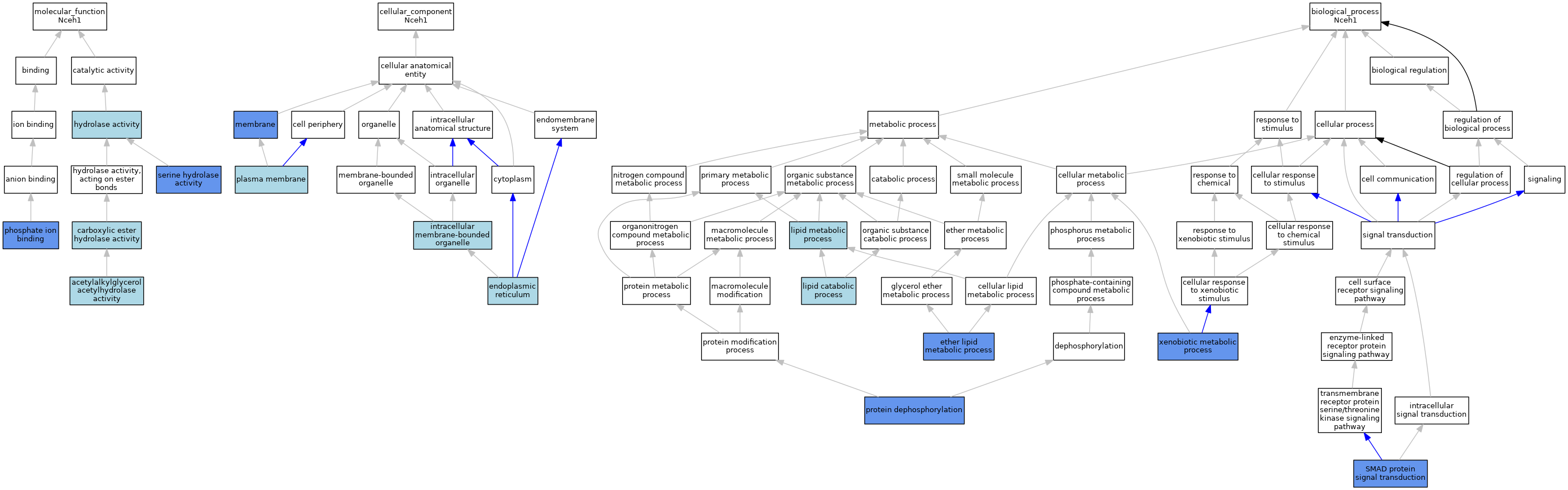
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0047378 | acetylalkylglycerol acetylhydrolase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0052689 | carboxylic ester hydrolase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042301 | phosphate ion binding | IDA | J:98213 | |||||||||
| Molecular Function | GO:0017171 | serine hydrolase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017171 | serine hydrolase activity | IDA | J:98213 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:98213 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0046485 | ether lipid metabolic process | IDA | J:165450 | |||||||||
| Biological Process | GO:0016042 | lipid catabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | IDA | J:98213 | |||||||||
| Biological Process | GO:0060395 | SMAD protein signal transduction | IDA | J:195995 | |||||||||
| Biological Process | GO:0006805 | xenobiotic metabolic process | IMP | J:98213 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||