|
Symbol Name ID |
Vash1
vasohibin 1 MGI:2442543 |
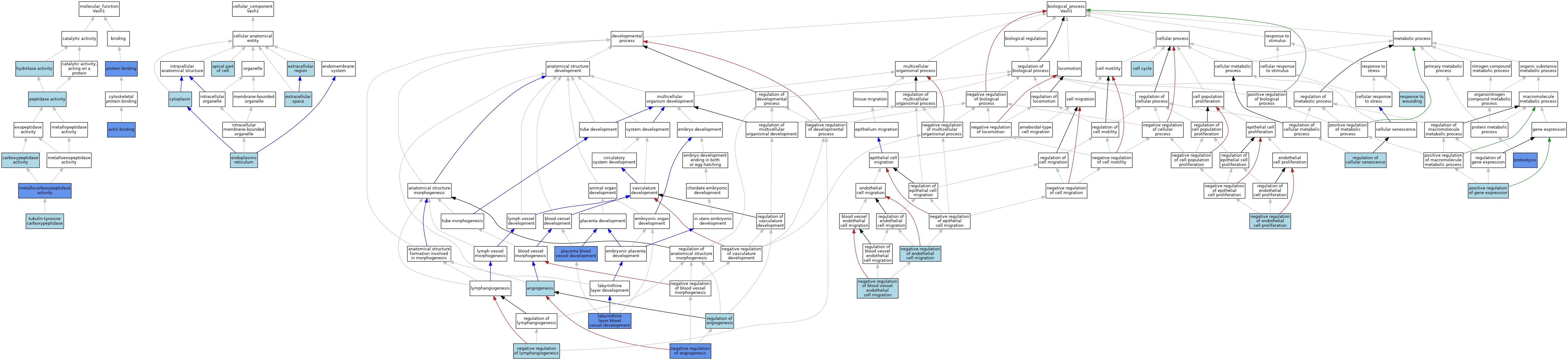
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003779 | actin binding | IDA | J:272424 | |||||||||
| Molecular Function | GO:0004180 | carboxypeptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004181 | metallocarboxypeptidase activity | IDA | J:272424 | |||||||||
| Molecular Function | GO:0004181 | metallocarboxypeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:272424 | |||||||||
| Molecular Function | GO:0106423 | tubulin-tyrosine carboxypeptidase | IEA | J:72245 | |||||||||
| Cellular Component | GO:0045177 | apical part of cell | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:93414 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:148726 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:73065 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | ISO | J:93414 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0060716 | labyrinthine layer blood vessel development | IMP | J:223264 | |||||||||
| Biological Process | GO:0016525 | negative regulation of angiogenesis | ISO | J:73065 | |||||||||
| Biological Process | GO:0016525 | negative regulation of angiogenesis | IDA | J:148726 | |||||||||
| Biological Process | GO:0016525 | negative regulation of angiogenesis | IMP | J:148726 | |||||||||
| Biological Process | GO:0043537 | negative regulation of blood vessel endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0010596 | negative regulation of endothelial cell migration | ISO | J:73065 | |||||||||
| Biological Process | GO:0001937 | negative regulation of endothelial cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0001937 | negative regulation of endothelial cell proliferation | ISO | J:148726 | |||||||||
| Biological Process | GO:1901491 | negative regulation of lymphangiogenesis | ISO | J:158639 | |||||||||
| Biological Process | GO:0060674 | placenta blood vessel development | IMP | J:223264 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | ISO | J:191937 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IDA | J:272424 | |||||||||
| Biological Process | GO:0006508 | proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0045765 | regulation of angiogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:2000772 | regulation of cellular senescence | ISO | J:191937 | |||||||||
| Biological Process | GO:0009611 | response to wounding | ISO | J:191937 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||