|
Symbol Name ID |
Gpr183
G protein-coupled receptor 183 MGI:2442034 |
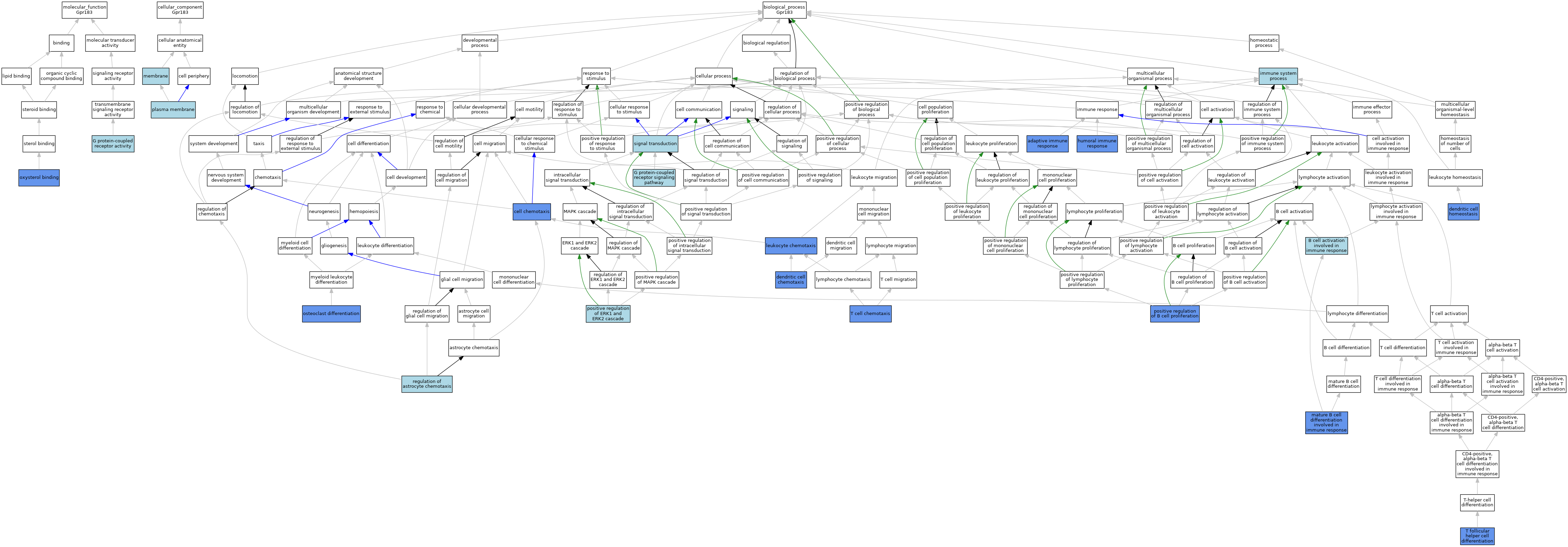
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008142 | oxysterol binding | IDA | J:174683 | |||||||||
| Molecular Function | GO:0008142 | oxysterol binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IMP | J:196453 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IMP | J:198681 | |||||||||
| Biological Process | GO:0002312 | B cell activation involved in immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0060326 | cell chemotaxis | IMP | J:229030 | |||||||||
| Biological Process | GO:0002407 | dendritic cell chemotaxis | IMP | J:196453 | |||||||||
| Biological Process | GO:0002407 | dendritic cell chemotaxis | IMP | J:198681 | |||||||||
| Biological Process | GO:0036145 | dendritic cell homeostasis | IMP | J:198681 | |||||||||
| Biological Process | GO:0036145 | dendritic cell homeostasis | IMP | J:196453 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0006959 | humoral immune response | IMP | J:151077 | |||||||||
| Biological Process | GO:0006959 | humoral immune response | IMP | J:198681 | |||||||||
| Biological Process | GO:0006959 | humoral immune response | IMP | J:196453 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0030595 | leukocyte chemotaxis | ISO | J:164563 | |||||||||
| Biological Process | GO:0030595 | leukocyte chemotaxis | IDA | J:219661 | |||||||||
| Biological Process | GO:0002313 | mature B cell differentiation involved in immune response | IMP | J:151077 | |||||||||
| Biological Process | GO:0030316 | osteoclast differentiation | IMP | J:229030 | |||||||||
| Biological Process | GO:0030890 | positive regulation of B cell proliferation | IDA | J:186017 | |||||||||
| Biological Process | GO:0030890 | positive regulation of B cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:2000458 | regulation of astrocyte chemotaxis | ISO | J:164563 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0010818 | T cell chemotaxis | IMP | J:233676 | |||||||||
| Biological Process | GO:0061470 | T follicular helper cell differentiation | IMP | J:233676 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||