|
Symbol Name ID |
Tpcn2
two pore segment channel 2 MGI:2385297 |
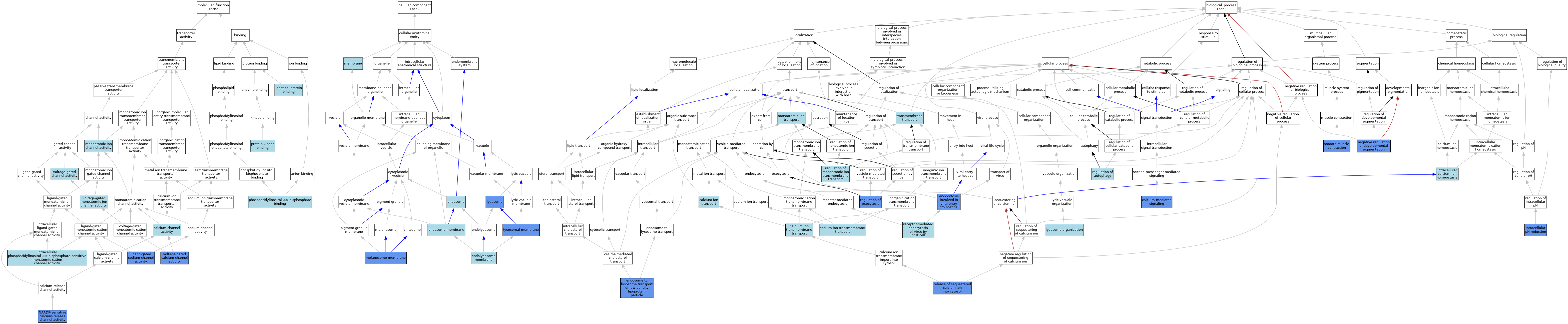
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005262 | calcium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0097682 | intracellular phosphatidylinositol-3,5-bisphosphate-sensitive monatomic cation channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0097682 | intracellular phosphatidylinositol-3,5-bisphosphate-sensitive monatomic cation channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | IDA | J:328828 | |||||||||
| Molecular Function | GO:0015280 | ligand-gated sodium channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005216 | monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0072345 | NAADP-sensitive calcium-release channel activity | IDA | J:175352 | |||||||||
| Molecular Function | GO:0072345 | NAADP-sensitive calcium-release channel activity | IMP | J:225206 | |||||||||
| Molecular Function | GO:0072345 | NAADP-sensitive calcium-release channel activity | IMP | J:175340 | |||||||||
| Molecular Function | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005245 | voltage-gated calcium channel activity | IMP | J:175340 | |||||||||
| Molecular Function | GO:0022832 | voltage-gated channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005244 | voltage-gated monoatomic ion channel activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0036020 | endolysosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0010008 | endosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005765 | lysosomal membrane | IDA | J:306599 | |||||||||
| Cellular Component | GO:0005765 | lysosomal membrane | IDA | J:175352 | |||||||||
| Cellular Component | GO:0005765 | lysosomal membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005765 | lysosomal membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IDA | J:175340 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IDA | J:225206 | |||||||||
| Cellular Component | GO:0005764 | lysosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0033162 | melanosome membrane | IDA | J:328828 | |||||||||
| Cellular Component | GO:0033162 | melanosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0070588 | calcium ion transmembrane transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0019722 | calcium-mediated signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0019722 | calcium-mediated signaling | IMP | J:225206 | |||||||||
| Biological Process | GO:0019722 | calcium-mediated signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0075509 | endocytosis involved in viral entry into host cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0075509 | endocytosis involved in viral entry into host cell | IMP | J:306587 | |||||||||
| Biological Process | GO:0075509 | endocytosis involved in viral entry into host cell | IBA | J:265628 | |||||||||
| Biological Process | GO:0090117 | endosome to lysosome transport of low-density lipoprotein particle | IMP | J:225206 | |||||||||
| Biological Process | GO:0006874 | intracellular calcium ion homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0051452 | intracellular pH reduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0051452 | intracellular pH reduction | IDA | J:328828 | |||||||||
| Biological Process | GO:0007040 | lysosome organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0048086 | negative regulation of developmental pigmentation | IDA | J:328828 | |||||||||
| Biological Process | GO:0048086 | negative regulation of developmental pigmentation | ISO | J:164563 | |||||||||
| Biological Process | GO:0019065 | receptor-mediated endocytosis of virus by host cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0010506 | regulation of autophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0017157 | regulation of exocytosis | IMP | J:306599 | |||||||||
| Biological Process | GO:0034765 | regulation of monoatomic ion transmembrane transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0051209 | release of sequestered calcium ion into cytosol | IMP | J:175340 | |||||||||
| Biological Process | GO:0051209 | release of sequestered calcium ion into cytosol | IMP | J:225206 | |||||||||
| Biological Process | GO:0006939 | smooth muscle contraction | IMP | J:165894 | |||||||||
| Biological Process | GO:0035725 | sodium ion transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0055085 | transmembrane transport | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||