|
Symbol Name ID |
Hkdc1
hexokinase domain containing 1 MGI:2384910 |
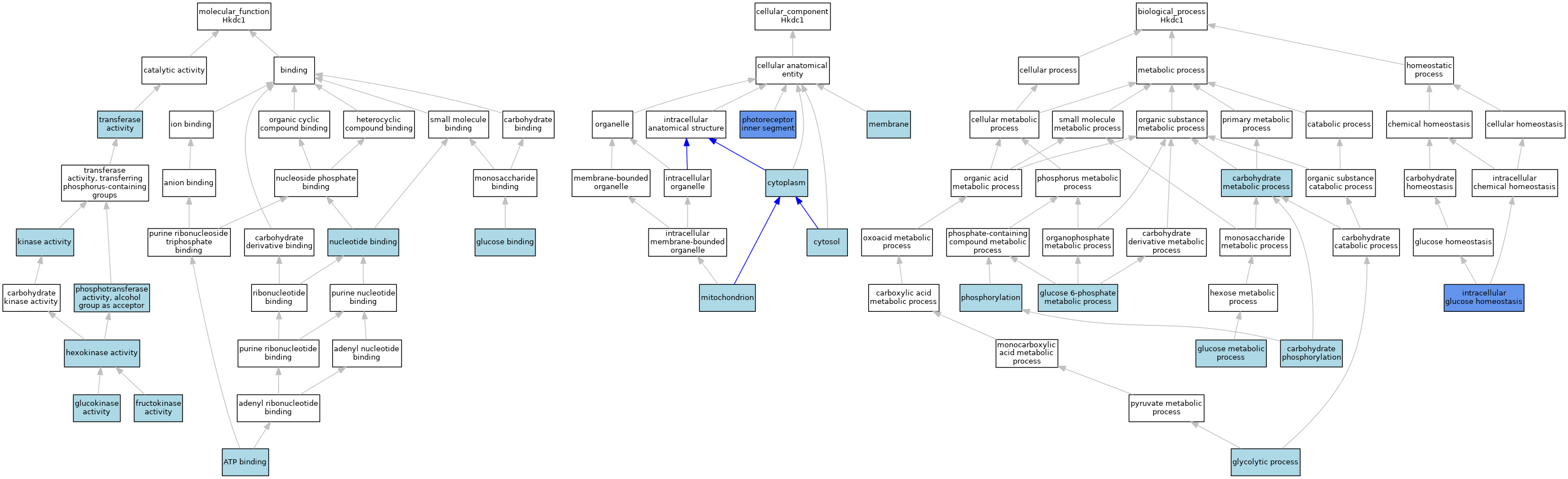
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0008865 | fructokinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004340 | glucokinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004340 | glucokinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005536 | glucose binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004396 | hexokinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004396 | hexokinase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016773 | phosphotransferase activity, alcohol group as acceptor | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0001917 | photoreceptor inner segment | IDA | J:266978 | |||||||||
| Biological Process | GO:0005975 | carbohydrate metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0046835 | carbohydrate phosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0051156 | glucose 6-phosphate metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051156 | glucose 6-phosphate metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006006 | glucose metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006096 | glycolytic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0001678 | intracellular glucose homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0001678 | intracellular glucose homeostasis | IMP | J:240354 | |||||||||
| Biological Process | GO:0001678 | intracellular glucose homeostasis | IMP | J:270636 | |||||||||
| Biological Process | GO:0001678 | intracellular glucose homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||