|
Symbol Name ID |
Ap1ar
adaptor-related protein complex 1 associated regulatory protein MGI:2384822 |
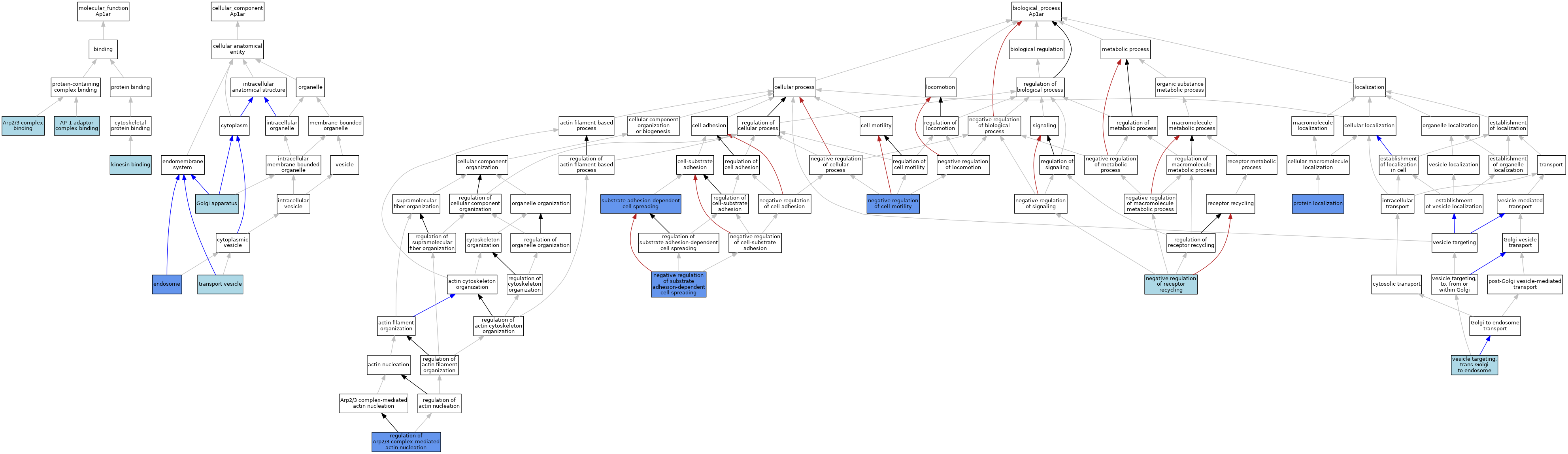
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0035650 | AP-1 adaptor complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035650 | AP-1 adaptor complex binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0035650 | AP-1 adaptor complex binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0071933 | Arp2/3 complex binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0019894 | kinesin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019894 | kinesin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019894 | kinesin binding | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005768 | endosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005768 | endosome | IDA | J:185589 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030133 | transport vesicle | ISO | J:164563 | |||||||||
| Biological Process | GO:2000146 | negative regulation of cell motility | IMP | J:185589 | |||||||||
| Biological Process | GO:0001920 | negative regulation of receptor recycling | ISO | J:164563 | |||||||||
| Biological Process | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | IMP | J:185589 | |||||||||
| Biological Process | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | IMP | J:185589 | |||||||||
| Biological Process | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | ISO | J:73065 | |||||||||
| Biological Process | GO:0008104 | protein localization | IMP | J:185589 | |||||||||
| Biological Process | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | IMP | J:185589 | |||||||||
| Biological Process | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | IGI | J:185589 | |||||||||
| Biological Process | GO:0034446 | substrate adhesion-dependent cell spreading | IMP | J:185589 | |||||||||
| Biological Process | GO:0048203 | vesicle targeting, trans-Golgi to endosome | ISO | J:164563 | |||||||||
| Biological Process | GO:0048203 | vesicle targeting, trans-Golgi to endosome | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||