|
Symbol Name ID |
Tnfrsf21
tumor necrosis factor receptor superfamily, member 21 MGI:2151075 |
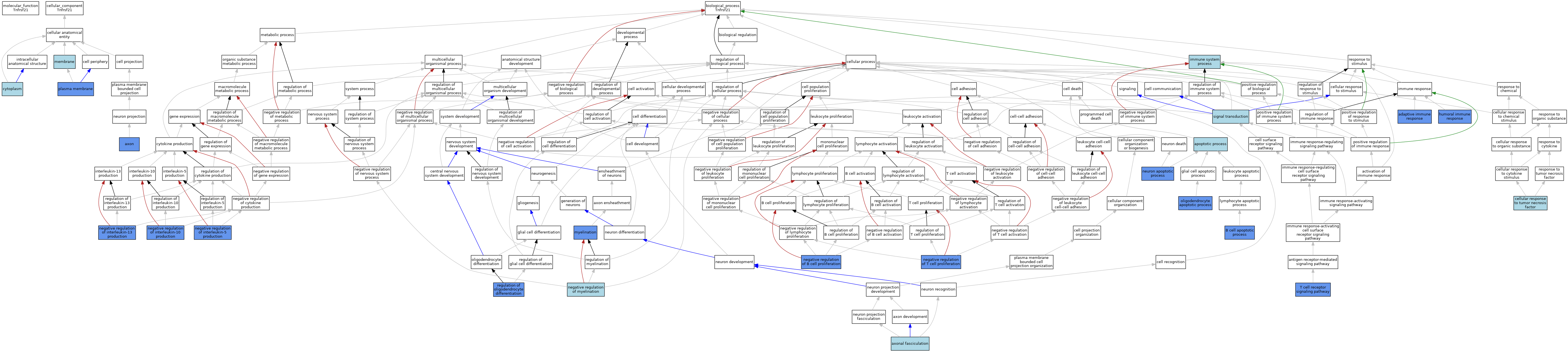
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular Component | GO:0030424 | axon | IDA | J:146602 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:109341 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IMP | J:70594 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0007413 | axonal fasciculation | ISO | J:155856 | |||||||||
| Biological Process | GO:0001783 | B cell apoptotic process | IMP | J:109341 | |||||||||
| Biological Process | GO:0071356 | cellular response to tumor necrosis factor | ISO | J:164563 | |||||||||
| Biological Process | GO:0006959 | humoral immune response | IMP | J:70594 | |||||||||
| Biological Process | GO:0006959 | humoral immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0042552 | myelination | IMP | J:205651 | |||||||||
| Biological Process | GO:0030889 | negative regulation of B cell proliferation | IMP | J:109341 | |||||||||
| Biological Process | GO:0030889 | negative regulation of B cell proliferation | IBA | J:265628 | |||||||||
| Biological Process | GO:0032693 | negative regulation of interleukin-10 production | IMP | J:70594 | |||||||||
| Biological Process | GO:0032696 | negative regulation of interleukin-13 production | IMP | J:70594 | |||||||||
| Biological Process | GO:0032714 | negative regulation of interleukin-5 production | IMP | J:70594 | |||||||||
| Biological Process | GO:0031642 | negative regulation of myelination | ISO | J:164563 | |||||||||
| Biological Process | GO:0031642 | negative regulation of myelination | IBA | J:265628 | |||||||||
| Biological Process | GO:0042130 | negative regulation of T cell proliferation | IMP | J:70594 | |||||||||
| Biological Process | GO:0042130 | negative regulation of T cell proliferation | IBA | J:265628 | |||||||||
| Biological Process | GO:0051402 | neuron apoptotic process | IMP | J:205745 | |||||||||
| Biological Process | GO:0051402 | neuron apoptotic process | IMP | J:146602 | |||||||||
| Biological Process | GO:0051402 | neuron apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0097252 | oligodendrocyte apoptotic process | IMP | J:205651 | |||||||||
| Biological Process | GO:0097252 | oligodendrocyte apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0048713 | regulation of oligodendrocyte differentiation | IMP | J:205651 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:0050852 | T cell receptor signaling pathway | IMP | J:70594 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||