|
Symbol Name ID |
Slc4a10
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 MGI:2150150 |
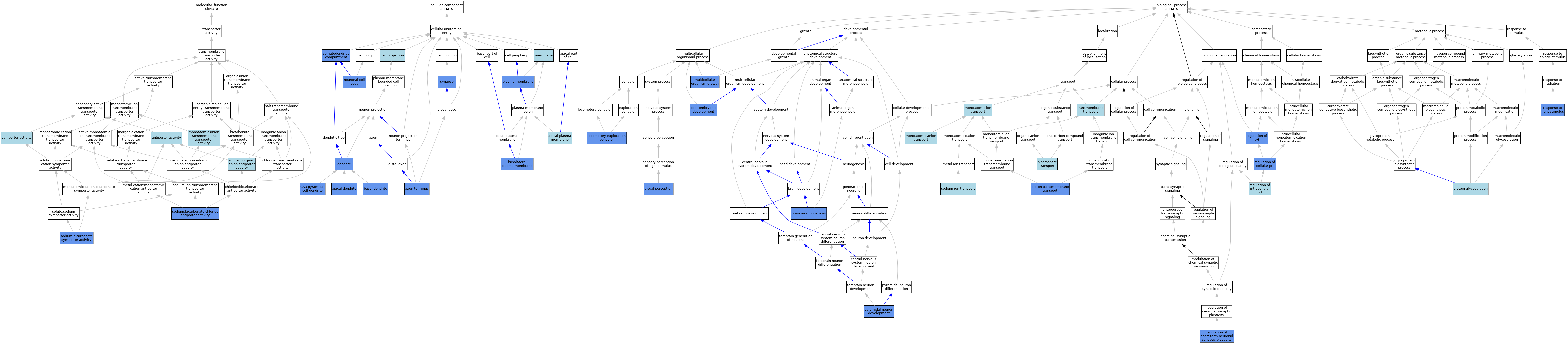
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0015297 | antiporter activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008509 | monoatomic anion transmembrane transporter activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0140892 | sodium,bicarbonate:chloride antiporter activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0140892 | sodium,bicarbonate:chloride antiporter activity | IDA | J:70983 | |||||||||
| Molecular Function | GO:0140892 | sodium,bicarbonate:chloride antiporter activity | IDA | J:273558 | |||||||||
| Molecular Function | GO:0008510 | sodium:bicarbonate symporter activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008510 | sodium:bicarbonate symporter activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008510 | sodium:bicarbonate symporter activity | IDA | J:273558 | |||||||||
| Molecular Function | GO:0008510 | sodium:bicarbonate symporter activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005452 | solute:inorganic anion antiporter activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0015293 | symporter activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0097440 | apical dendrite | IDA | J:131058 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043679 | axon terminus | IDA | J:191940 | |||||||||
| Cellular Component | GO:0097441 | basal dendrite | IDA | J:131058 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | IDA | J:130810 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | IDA | J:271710 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | IDA | J:176751 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | IDA | J:87933 | |||||||||
| Cellular Component | GO:0097442 | CA3 pyramidal cell dendrite | IDA | J:131058 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:274854 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:191940 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:191940 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:131058 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:201360 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISA | J:70983 | |||||||||
| Cellular Component | GO:0036477 | somatodendritic compartment | IDA | J:274854 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:239653 | |||||||||
| Biological Process | GO:0015701 | bicarbonate transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0048854 | brain morphogenesis | IMP | J:131058 | |||||||||
| Biological Process | GO:0035641 | locomotory exploration behavior | IMP | J:131058 | |||||||||
| Biological Process | GO:0006820 | monoatomic anion transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0035264 | multicellular organism growth | IMP | J:131058 | |||||||||
| Biological Process | GO:0009791 | post-embryonic development | IMP | J:131058 | |||||||||
| Biological Process | GO:0006486 | protein glycosylation | ISO | J:87933 | |||||||||
| Biological Process | GO:1902600 | proton transmembrane transport | IMP | J:131058 | |||||||||
| Biological Process | GO:0021860 | pyramidal neuron development | IMP | J:131058 | |||||||||
| Biological Process | GO:0030641 | regulation of cellular pH | IMP | J:131058 | |||||||||
| Biological Process | GO:0051453 | regulation of intracellular pH | IBA | J:265628 | |||||||||
| Biological Process | GO:0006885 | regulation of pH | IDA | J:70983 | |||||||||
| Biological Process | GO:0048172 | regulation of short-term neuronal synaptic plasticity | IMP | J:239653 | |||||||||
| Biological Process | GO:0009416 | response to light stimulus | IMP | J:131058 | |||||||||
| Biological Process | GO:0006814 | sodium ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0055085 | transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0007601 | visual perception | IMP | J:191940 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||