|
Symbol Name ID |
Spred2
sprouty-related EVH1 domain containing 2 MGI:2150019 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
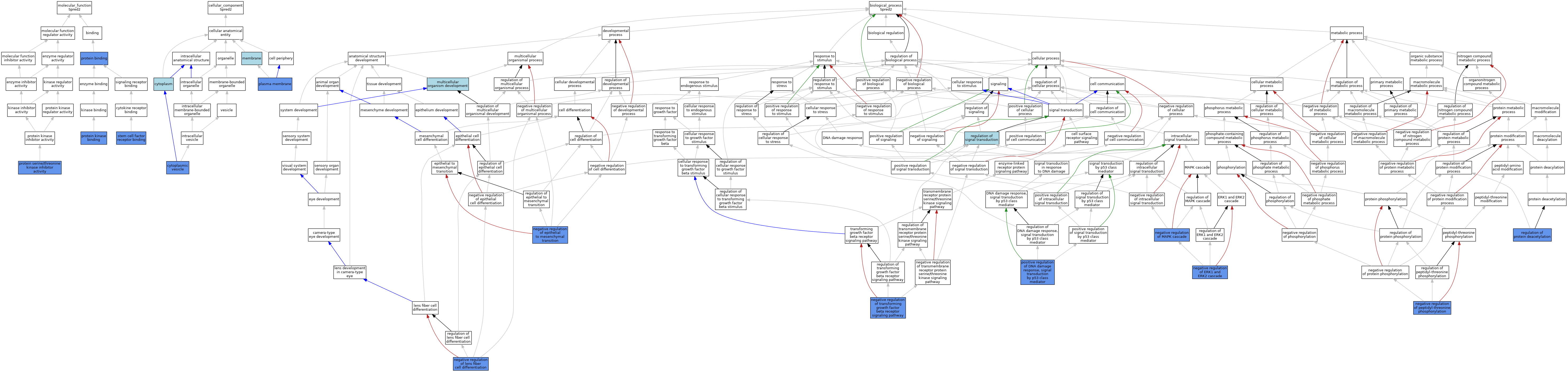
| Molecular Function | GO:0005515 | protein binding | IPI | J:153806 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:130605 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:82566 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IPI | J:183209 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0030291 | protein serine/threonine kinase inhibitor activity | IDA | J:183209 | |||||||||
| Molecular Function | GO:0005173 | stem cell factor receptor binding | IPI | J:82566 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IDA | J:108170 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82566 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:72247 | |||||||||
| Biological Process | GO:0010719 | negative regulation of epithelial to mesenchymal transition | IDA | J:287032 | |||||||||
| Biological Process | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | IDA | J:287039 | |||||||||
| Biological Process | GO:1902747 | negative regulation of lens fiber cell differentiation | IDA | J:287039 | |||||||||
| Biological Process | GO:0043409 | negative regulation of MAPK cascade | IBA | J:265628 | |||||||||
| Biological Process | GO:0043409 | negative regulation of MAPK cascade | IMP | J:82566 | |||||||||
| Biological Process | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | IDA | J:183209 | |||||||||
| Biological Process | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | IDA | J:287032 | |||||||||
| Biological Process | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | IDA | J:183209 | |||||||||
| Biological Process | GO:0090311 | regulation of protein deacetylation | IDA | J:183209 | |||||||||
| Biological Process | GO:0009966 | regulation of signal transduction | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||