|
Symbol Name ID |
Ffar4
free fatty acid receptor 4 MGI:2147577 |
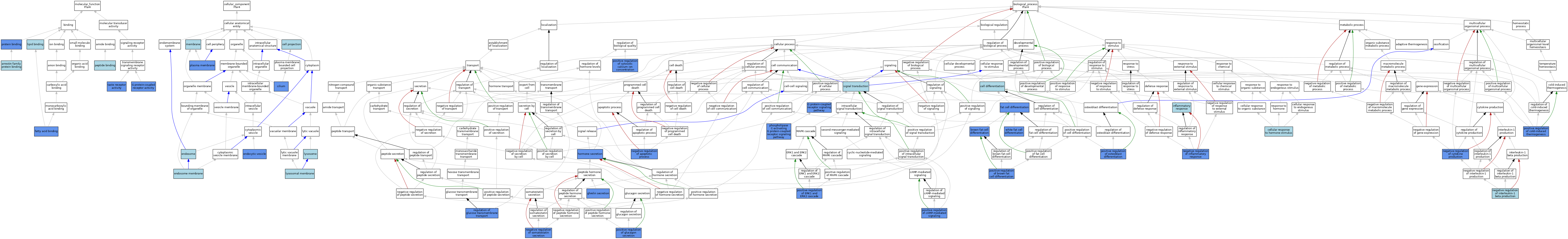
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:1990763 | arrestin family protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005504 | fatty acid binding | IDA | J:166091 | |||||||||
| Molecular Function | GO:0005504 | fatty acid binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005504 | fatty acid binding | IDA | J:95456 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IDA | J:95456 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042277 | peptide binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:166091 | |||||||||
| Molecular Function | GO:0008527 | taste receptor activity | IMP | J:161517 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005929 | cilium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005929 | cilium | IDA | J:282613 | |||||||||
| Cellular Component | GO:0030139 | endocytic vesicle | IDA | J:95456 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0010008 | endosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005765 | lysosomal membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:166091 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-444204 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:95456 | |||||||||
| Biological Process | GO:0050873 | brown fat cell differentiation | IMP | J:240649 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0032870 | cellular response to hormone stimulus | IBA | J:265628 | |||||||||
| Biological Process | GO:0045444 | fat cell differentiation | IMP | J:182599 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IDA | J:95456 | |||||||||
| Biological Process | GO:0036321 | ghrelin secretion | IMP | J:282608 | |||||||||
| Biological Process | GO:0046879 | hormone secretion | IBA | J:265628 | |||||||||
| Biological Process | GO:0046879 | hormone secretion | IDA | J:95456 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IEA | J:60000 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IMP | J:99416 | |||||||||
| Biological Process | GO:0001818 | negative regulation of cytokine production | IDA | J:166091 | |||||||||
| Biological Process | GO:0001818 | negative regulation of cytokine production | IMP | J:232433 | |||||||||
| Biological Process | GO:0001818 | negative regulation of cytokine production | IMP | J:232433 | |||||||||
| Biological Process | GO:0050728 | negative regulation of inflammatory response | IDA | J:166091 | |||||||||
| Biological Process | GO:0032691 | negative regulation of interleukin-1 beta production | ISO | J:164563 | |||||||||
| Biological Process | GO:0090275 | negative regulation of somatostatin secretion | IMP | J:211675 | |||||||||
| Biological Process | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | IMP | J:283203 | |||||||||
| Biological Process | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0090336 | positive regulation of brown fat cell differentiation | IMP | J:261351 | |||||||||
| Biological Process | GO:0043950 | positive regulation of cAMP-mediated signaling | IDA | J:282613 | |||||||||
| Biological Process | GO:0120162 | positive regulation of cold-induced thermogenesis | IMP | J:240649 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | ISO | J:164563 | |||||||||
| Biological Process | GO:0007204 | positive regulation of cytosolic calcium ion concentration | IMP | J:283203 | |||||||||
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | IMP | J:99416 | |||||||||
| Biological Process | GO:0070094 | positive regulation of glucagon secretion | IMP | J:214106 | |||||||||
| Biological Process | GO:0045669 | positive regulation of osteoblast differentiation | IMP | J:283205 | |||||||||
| Biological Process | GO:0010827 | regulation of glucose transmembrane transport | IDA | J:166091 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0050872 | white fat cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0050872 | white fat cell differentiation | IDA | J:282613 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||