|
Symbol Name ID |
Dhx29
DExH-box helicase 29 MGI:2145374 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
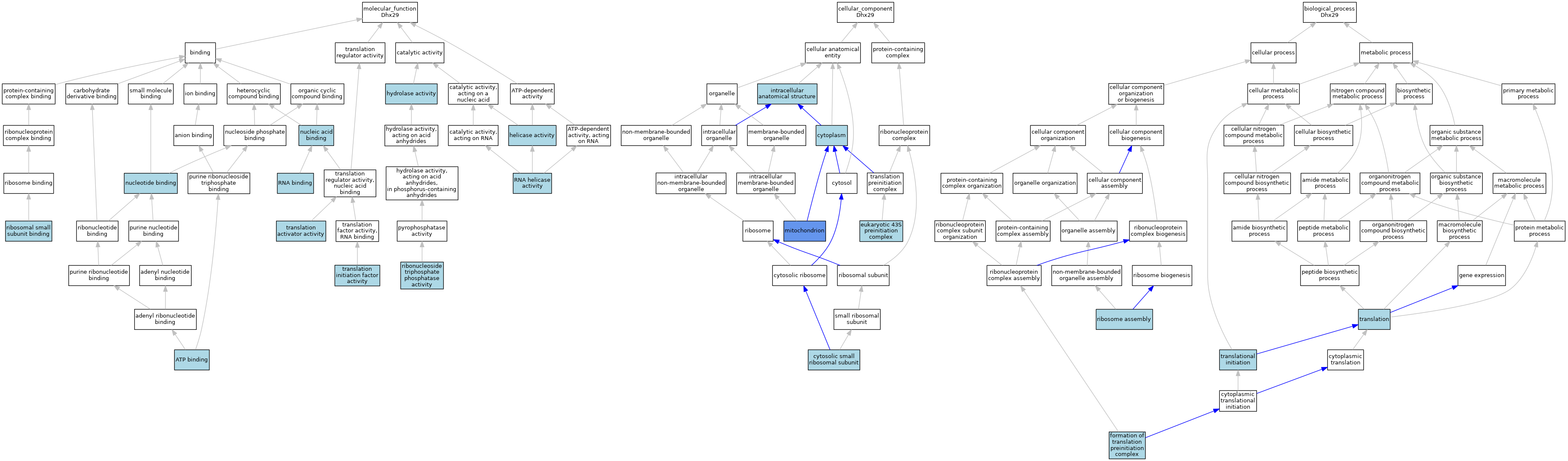
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004386 | helicase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0017111 | ribonucleoside triphosphate phosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043024 | ribosomal small subunit binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003724 | RNA helicase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003724 | RNA helicase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0008494 | translation activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003743 | translation initiation factor activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0022627 | cytosolic small ribosomal subunit | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016282 | eukaryotic 43S preinitiation complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005622 | intracellular anatomical structure | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Biological Process | GO:0001731 | formation of translation preinitiation complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0042255 | ribosome assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0006412 | translation | IEA | J:60000 | |||||||||
| Biological Process | GO:0006413 | translational initiation | IEA | J:72247 | |||||||||
| Biological Process | GO:0006413 | translational initiation | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||