|
Symbol Name ID |
Dpyd
dihydropyrimidine dehydrogenase MGI:2139667 |
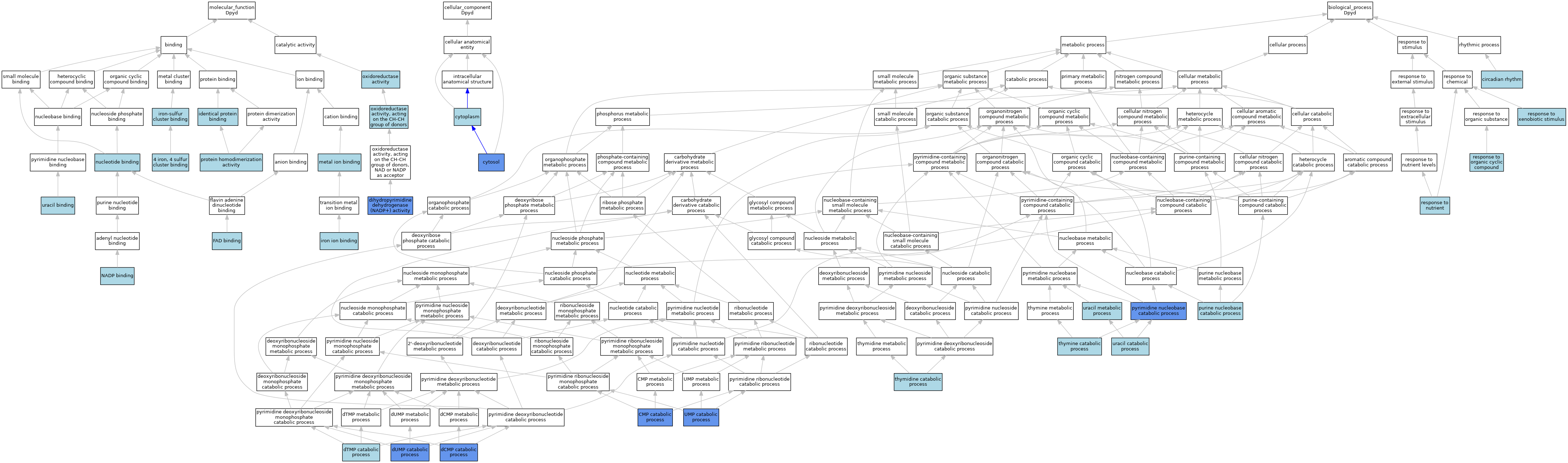
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0051539 | 4 iron, 4 sulfur cluster binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | IDA | J:5178 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | IDA | J:5178 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | ISO | J:21749 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | IDA | J:5178 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | IDA | J:5178 | |||||||||
| Molecular Function | GO:0017113 | dihydropyrimidine dehydrogenase (NADP+) activity | IDA | J:83143 | |||||||||
| Molecular Function | GO:0071949 | FAD binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005506 | iron ion binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0051536 | iron-sulfur cluster binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051536 | iron-sulfur cluster binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0050661 | NADP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0050661 | NADP binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors | IEA | J:72247 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0002058 | uracil binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0002058 | uracil binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:5178 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:5178 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:83143 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:83143 | |||||||||
| Biological Process | GO:0007623 | circadian rhythm | ISO | J:155856 | |||||||||
| Biological Process | GO:0006248 | CMP catabolic process | IDA | J:5178 | |||||||||
| Biological Process | GO:0006249 | dCMP catabolic process | IDA | J:5178 | |||||||||
| Biological Process | GO:0046074 | dTMP catabolic process | ISO | J:21749 | |||||||||
| Biological Process | GO:0046079 | dUMP catabolic process | IDA | J:5178 | |||||||||
| Biological Process | GO:0006145 | purine nucleobase catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006208 | pyrimidine nucleobase catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006208 | pyrimidine nucleobase catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006208 | pyrimidine nucleobase catabolic process | IDA | J:83143 | |||||||||
| Biological Process | GO:0007584 | response to nutrient | ISO | J:155856 | |||||||||
| Biological Process | GO:0014070 | response to organic cyclic compound | ISO | J:155856 | |||||||||
| Biological Process | GO:0009410 | response to xenobiotic stimulus | ISO | J:155856 | |||||||||
| Biological Process | GO:0006214 | thymidine catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006210 | thymine catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006210 | thymine catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0046050 | UMP catabolic process | IDA | J:5178 | |||||||||
| Biological Process | GO:0006212 | uracil catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006212 | uracil catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0019860 | uracil metabolic process | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||