|
Symbol Name ID |
Grin3a
glutamate receptor ionotropic, NMDA3A MGI:1933206 |
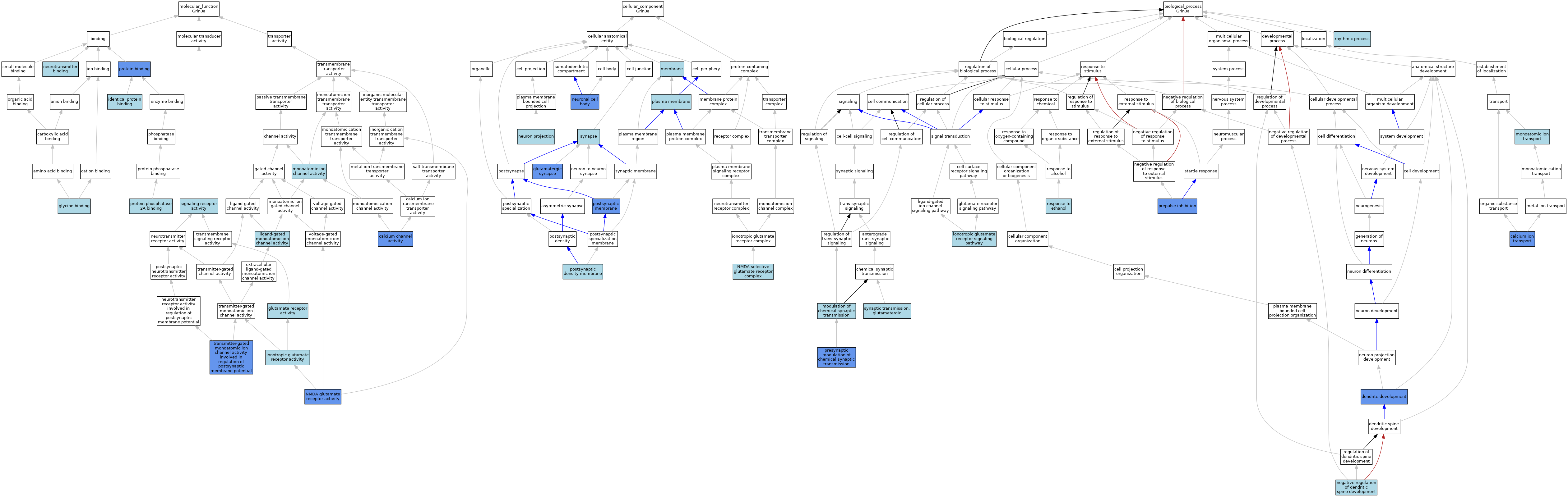
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005262 | calcium channel activity | IDA | J:66481 | |||||||||
| Molecular Function | GO:0005262 | calcium channel activity | IMP | J:85752 | |||||||||
| Molecular Function | GO:0008066 | glutamate receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016594 | glycine binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016594 | glycine binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004970 | ionotropic glutamate receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015276 | ligand-gated monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005216 | monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0042165 | neurotransmitter binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004972 | NMDA glutamate receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004972 | NMDA glutamate receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004972 | NMDA glutamate receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004972 | NMDA glutamate receptor activity | IMP | J:85752 | |||||||||
| Molecular Function | GO:0004972 | NMDA glutamate receptor activity | IPI | J:66481 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:205503 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:66481 | |||||||||
| Molecular Function | GO:0051721 | protein phosphatase 2A binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0038023 | signaling receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:1904315 | transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential | IBA | J:265628 | |||||||||
| Molecular Function | GO:1904315 | transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential | IDA | J:265614 | |||||||||
| Molecular Function | GO:1904315 | transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential | IMP | J:265614 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IMP | J:253352 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:253352 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:265614 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IMP | J:265614 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:85752 | |||||||||
| Cellular Component | GO:0017146 | NMDA selective glutamate receptor complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0017146 | NMDA selective glutamate receptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0098839 | postsynaptic density membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098839 | postsynaptic density membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | IDA | J:66481 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:164563 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006816 | calcium ion transport | IMP | J:85752 | |||||||||
| Biological Process | GO:0016358 | dendrite development | IMP | J:66481 | |||||||||
| Biological Process | GO:0035235 | ionotropic glutamate receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0035235 | ionotropic glutamate receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0050804 | modulation of chemical synaptic transmission | IBA | J:265628 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0061000 | negative regulation of dendritic spine development | ISO | J:155856 | |||||||||
| Biological Process | GO:0060134 | prepulse inhibition | IMP | J:102120 | |||||||||
| Biological Process | GO:0099171 | presynaptic modulation of chemical synaptic transmission | IDA | J:253352 | |||||||||
| Biological Process | GO:0099171 | presynaptic modulation of chemical synaptic transmission | IMP | J:253352 | |||||||||
| Biological Process | GO:0045471 | response to ethanol | ISO | J:164563 | |||||||||
| Biological Process | GO:0048511 | rhythmic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0035249 | synaptic transmission, glutamatergic | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||