|
Symbol Name ID |
Sirt3
sirtuin 3 MGI:1927665 |
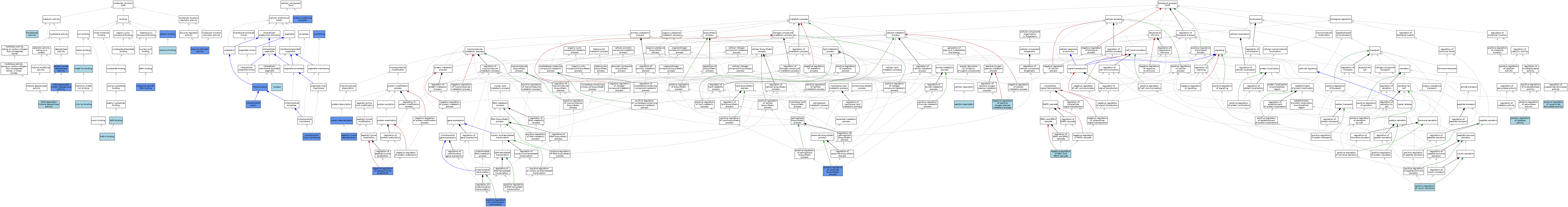
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008047 | enzyme activator activity | IDA | J:176493 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051287 | NAD binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0070403 | NAD+ binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017136 | NAD-dependent histone deacetylase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0034979 | NAD-dependent protein deacetylase activity | IDA | J:230048 | |||||||||
| Molecular Function | GO:0034979 | NAD-dependent protein deacetylase activity | IDA | J:276379 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:230048 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:257325 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:174774 | |||||||||
| Molecular Function | GO:0033558 | protein lysine deacetylase activity | IDA | J:176493 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | IDA | J:257325 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:65632 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | IDA | J:130436 | |||||||||
| Cellular Component | GO:0005759 | mitochondrial matrix | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005759 | mitochondrial matrix | IDA | J:129009 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:129009 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:233904 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | IDA | J:257325 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0009060 | aerobic respiration | ISO | J:164563 | |||||||||
| Biological Process | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | ISO | J:155856 | |||||||||
| Biological Process | GO:2000757 | negative regulation of peptidyl-lysine acetylation | IDA | J:129009 | |||||||||
| Biological Process | GO:2000757 | negative regulation of peptidyl-lysine acetylation | IMP | J:129009 | |||||||||
| Biological Process | GO:2000378 | negative regulation of reactive oxygen species metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0034983 | peptidyl-lysine deacetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0034983 | peptidyl-lysine deacetylation | IDA | J:230048 | |||||||||
| Biological Process | GO:1902553 | positive regulation of catalase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:2000304 | positive regulation of ceramide biosynthetic process | IDA | J:230048 | |||||||||
| Biological Process | GO:0032024 | positive regulation of insulin secretion | ISO | J:155856 | |||||||||
| Biological Process | GO:1903109 | positive regulation of mitochondrial transcription | IMP | J:257325 | |||||||||
| Biological Process | GO:1901671 | positive regulation of superoxide dismutase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0006476 | protein deacetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006476 | protein deacetylation | IMP | J:196142 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||