|
Symbol Name ID |
Ddrgk1
DDRGK domain containing 1 MGI:1924256 |
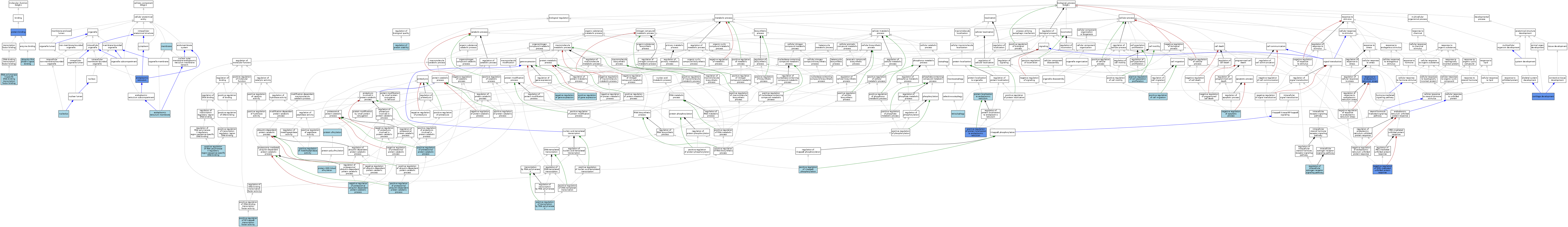
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | IPI | J:217277 | |||||||||
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0044389 | ubiquitin-like protein ligase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:217277 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:73065 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Biological Process | GO:0051216 | cartilage development | IMP | J:243374 | |||||||||
| Biological Process | GO:0051216 | cartilage development | ISO | J:164563 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response | IMP | J:244198 | |||||||||
| Biological Process | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response | ISO | J:164563 | |||||||||
| Biological Process | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:1903721 | positive regulation of I-kappaB phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:1905050 | positive regulation of metallopeptidase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:1901800 | positive regulation of proteasomal protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:1905552 | positive regulation of protein localization to endoplasmic reticulum | IDA | J:217277 | |||||||||
| Biological Process | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:1990592 | protein K69-linked ufmylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0070972 | protein localization to endoplasmic reticulum | ISO | J:164563 | |||||||||
| Biological Process | GO:0071569 | protein ufmylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0031647 | regulation of protein stability | ISO | J:164563 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | IMP | J:217277 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | ISO | J:164563 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | IMP | J:244198 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||