|
Symbol Name ID |
C2cd5
C2 calcium-dependent domain containing 5 MGI:1921991 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
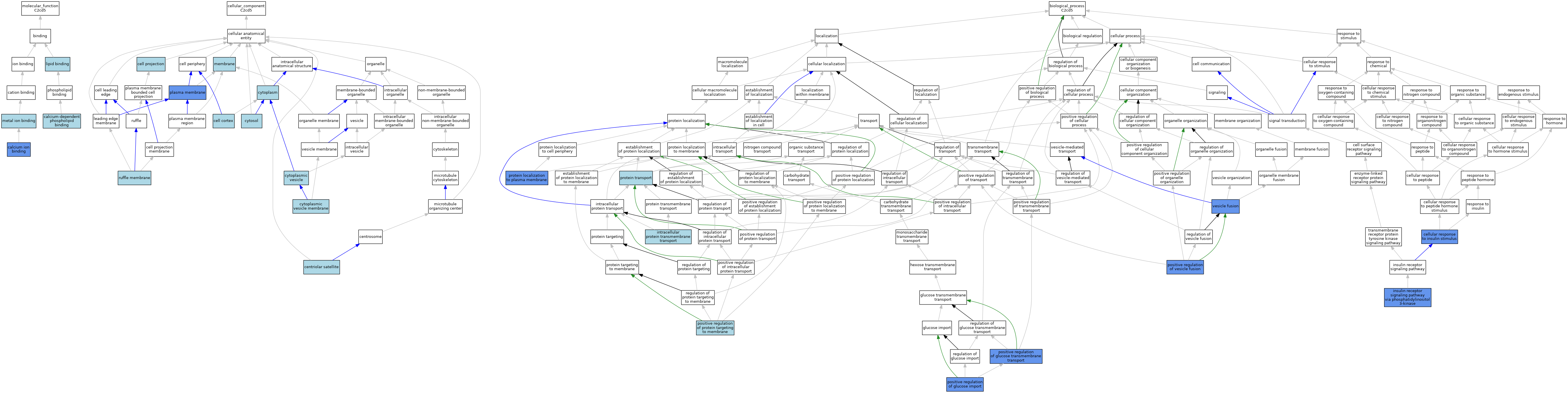
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005509 | calcium ion binding | IDA | J:176652 | |||||||||
| Molecular Function | GO:0005544 | calcium-dependent phospholipid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005544 | calcium-dependent phospholipid binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005938 | cell cortex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0034451 | centriolar satellite | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030659 | cytoplasmic vesicle membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-5263624 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:176652 | |||||||||
| Cellular Component | GO:0032587 | ruffle membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0032869 | cellular response to insulin stimulus | IDA | J:176652 | |||||||||
| Biological Process | GO:0032869 | cellular response to insulin stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | IMP | J:176652 | |||||||||
| Biological Process | GO:0065002 | intracellular protein transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0065002 | intracellular protein transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0046326 | positive regulation of glucose import | IGI | J:176652 | |||||||||
| Biological Process | GO:0046326 | positive regulation of glucose import | IMP | J:176652 | |||||||||
| Biological Process | GO:0010828 | positive regulation of glucose transmembrane transport | IMP | J:176652 | |||||||||
| Biological Process | GO:0010828 | positive regulation of glucose transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0090314 | positive regulation of protein targeting to membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0090314 | positive regulation of protein targeting to membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0031340 | positive regulation of vesicle fusion | IMP | J:176652 | |||||||||
| Biological Process | GO:0031340 | positive regulation of vesicle fusion | ISO | J:164563 | |||||||||
| Biological Process | GO:0031340 | positive regulation of vesicle fusion | IBA | J:265628 | |||||||||
| Biological Process | GO:0072659 | protein localization to plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0072659 | protein localization to plasma membrane | IGI | J:176652 | |||||||||
| Biological Process | GO:0072659 | protein localization to plasma membrane | IMP | J:176652 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006906 | vesicle fusion | IMP | J:176652 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||