|
Symbol Name ID |
Setd7
SET domain containing (lysine methyltransferase) 7 MGI:1920501 |
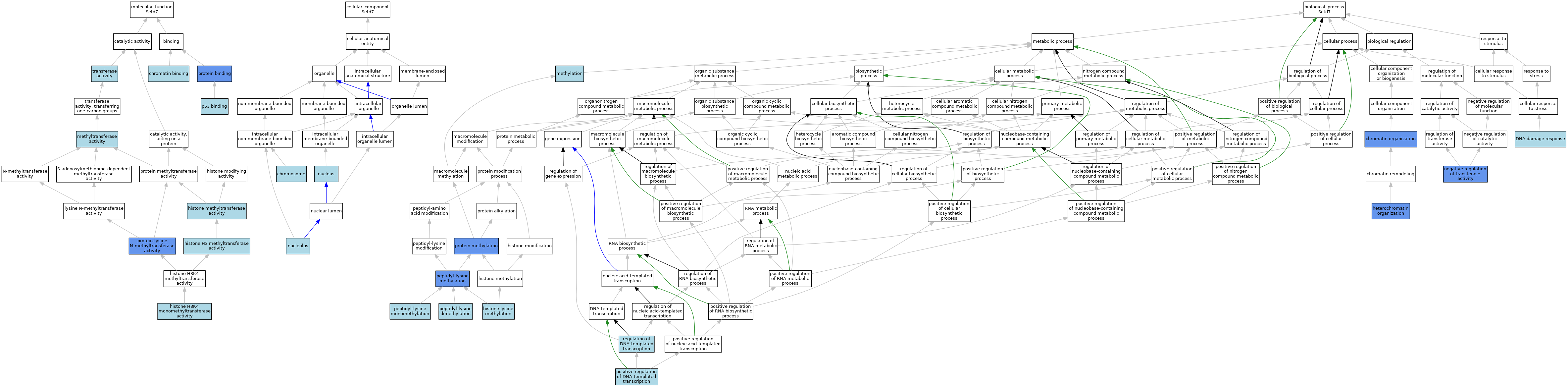
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0140938 | histone H3 methyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140945 | histone H3K4 monomethyltransferase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0042054 | histone methyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008168 | methyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0002039 | p53 binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:194231 | |||||||||
| Molecular Function | GO:0016279 | protein-lysine N-methyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016279 | protein-lysine N-methyltransferase activity | IDA | J:238299 | |||||||||
| Molecular Function | GO:0016279 | protein-lysine N-methyltransferase activity | IDA | J:326061 | |||||||||
| Molecular Function | GO:0016279 | protein-lysine N-methyltransferase activity | IDA | J:194231 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005694 | chromosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | IDA | J:194231 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:194231 | |||||||||
| Biological Process | GO:0070828 | heterochromatin organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0070828 | heterochromatin organization | ISO | J:194231 | |||||||||
| Biological Process | GO:0070828 | heterochromatin organization | IMP | J:194231 | |||||||||
| Biological Process | GO:0034968 | histone lysine methylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0032259 | methylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0051348 | negative regulation of transferase activity | IDA | J:194231 | |||||||||
| Biological Process | GO:0018027 | peptidyl-lysine dimethylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0018022 | peptidyl-lysine methylation | IDA | J:326061 | |||||||||
| Biological Process | GO:0018022 | peptidyl-lysine methylation | IDA | J:194231 | |||||||||
| Biological Process | GO:0018026 | peptidyl-lysine monomethylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0018026 | peptidyl-lysine monomethylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:155856 | |||||||||
| Biological Process | GO:0006479 | protein methylation | IDA | J:238299 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||