|
Symbol Name ID |
Prcp
prolylcarboxypeptidase (angiotensinase C) MGI:1919711 |
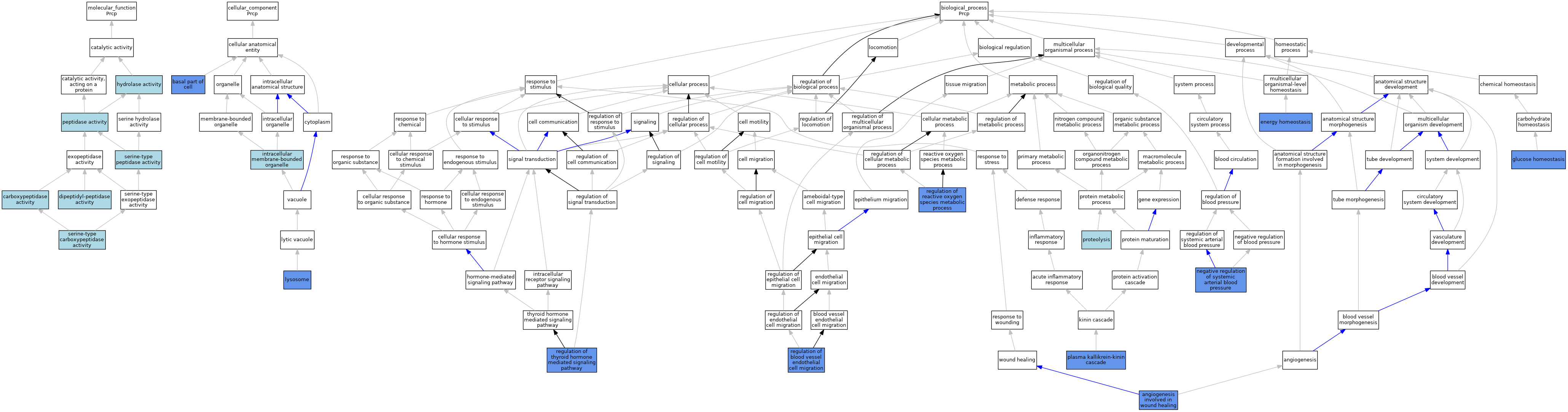
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004180 | carboxypeptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008239 | dipeptidyl-peptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004185 | serine-type carboxypeptidase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0008236 | serine-type peptidase activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0045178 | basal part of cell | IDA | J:172848 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IDA | J:261411 | |||||||||
| Biological Process | GO:0060055 | angiogenesis involved in wound healing | IBA | J:265628 | |||||||||
| Biological Process | GO:0060055 | angiogenesis involved in wound healing | IMP | J:201780 | |||||||||
| Biological Process | GO:0097009 | energy homeostasis | IMP | J:181710 | |||||||||
| Biological Process | GO:0097009 | energy homeostasis | IMP | J:187028 | |||||||||
| Biological Process | GO:0042593 | glucose homeostasis | IMP | J:187028 | |||||||||
| Biological Process | GO:0003085 | negative regulation of systemic arterial blood pressure | IBA | J:265628 | |||||||||
| Biological Process | GO:0003085 | negative regulation of systemic arterial blood pressure | IMP | J:172848 | |||||||||
| Biological Process | GO:0002353 | plasma kallikrein-kinin cascade | IMP | J:172848 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:72247 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0043535 | regulation of blood vessel endothelial cell migration | IBA | J:265628 | |||||||||
| Biological Process | GO:0043535 | regulation of blood vessel endothelial cell migration | ISO | J:73065 | |||||||||
| Biological Process | GO:0043535 | regulation of blood vessel endothelial cell migration | IMP | J:201780 | |||||||||
| Biological Process | GO:2000377 | regulation of reactive oxygen species metabolic process | IMP | J:172848 | |||||||||
| Biological Process | GO:0002155 | regulation of thyroid hormone mediated signaling pathway | IMP | J:181710 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||