|
Symbol Name ID |
Spink5
serine peptidase inhibitor, Kazal type 5 MGI:1919682 |
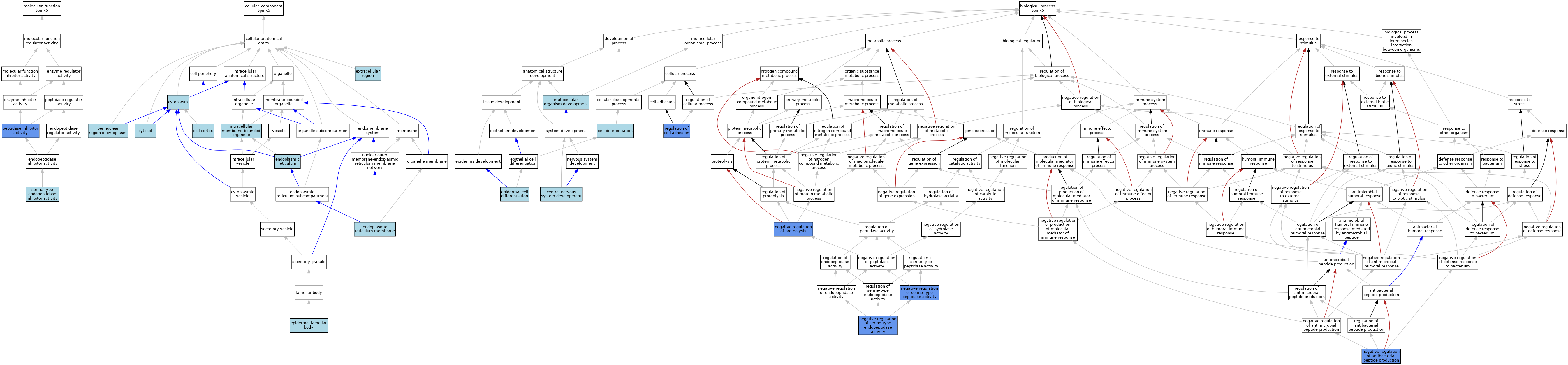
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0030414 | peptidase inhibitor activity | IMP | J:93050 | |||||||||
| Molecular Function | GO:0004867 | serine-type endopeptidase inhibitor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004867 | serine-type endopeptidase inhibitor activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005938 | cell cortex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0097209 | epidermal lamellar body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0007417 | central nervous system development | IBA | J:265628 | |||||||||
| Biological Process | GO:0009913 | epidermal cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IBA | J:265628 | |||||||||
| Biological Process | GO:0002787 | negative regulation of antibacterial peptide production | IMP | J:129736 | |||||||||
| Biological Process | GO:0045861 | negative regulation of proteolysis | IMP | J:93050 | |||||||||
| Biological Process | GO:1900004 | negative regulation of serine-type endopeptidase activity | IMP | J:129736 | |||||||||
| Biological Process | GO:1902572 | negative regulation of serine-type peptidase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:1902572 | negative regulation of serine-type peptidase activity | IMP | J:158765 | |||||||||
| Biological Process | GO:0030155 | regulation of cell adhesion | IMP | J:93050 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||