|
Symbol Name ID |
Seh1l
SEH1-like (S. cerevisiae MGI:1919374 |
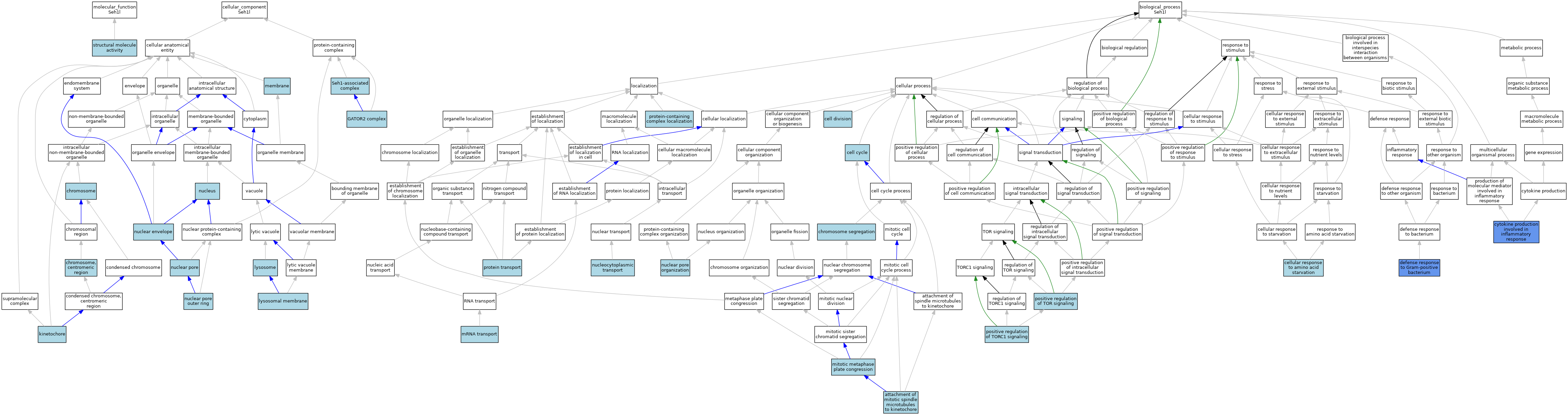
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005198 | structural molecule activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0000775 | chromosome, centromeric region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0061700 | GATOR2 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005765 | lysosomal membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005635 | nuclear envelope | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005643 | nuclear pore | NAS | J:320054 | |||||||||
| Cellular Component | GO:0031080 | nuclear pore outer ring | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031080 | nuclear pore outer ring | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0035859 | Seh1-associated complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | ISO | J:164563 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0051301 | cell division | IEA | J:60000 | |||||||||
| Biological Process | GO:0034198 | cellular response to amino acid starvation | ISO | J:164563 | |||||||||
| Biological Process | GO:0034198 | cellular response to amino acid starvation | IBA | J:265628 | |||||||||
| Biological Process | GO:0007059 | chromosome segregation | IEA | J:60000 | |||||||||
| Biological Process | GO:0002534 | cytokine production involved in inflammatory response | IMP | J:163990 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IMP | J:163990 | |||||||||
| Biological Process | GO:0007080 | mitotic metaphase plate congression | ISO | J:164563 | |||||||||
| Biological Process | GO:0051028 | mRNA transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006999 | nuclear pore organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0006913 | nucleocytoplasmic transport | NAS | J:320054 | |||||||||
| Biological Process | GO:0032008 | positive regulation of TOR signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:1904263 | positive regulation of TORC1 signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:1904263 | positive regulation of TORC1 signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0031503 | protein-containing complex localization | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||