|
Symbol Name ID |
Sash1
SAM and SH3 domain containing 1 MGI:1917347 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
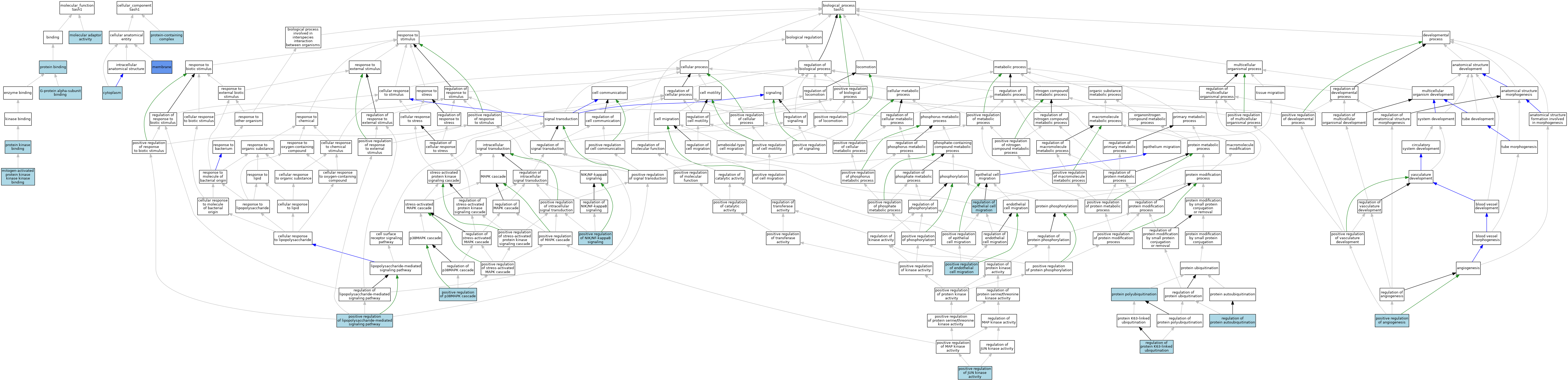
| Molecular Function | GO:0001965 | G-protein alpha-subunit binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031435 | mitogen-activated protein kinase kinase kinase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031435 | mitogen-activated protein kinase kinase kinase binding | ISO | J:205449 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | ISO | J:205449 | |||||||||
| Molecular Function | GO:0005515 | protein binding | ISO | J:205449 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:205449 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:205449 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:205449 | |||||||||
| Biological Process | GO:0045766 | positive regulation of angiogenesis | ISO | J:205449 | |||||||||
| Biological Process | GO:0010595 | positive regulation of endothelial cell migration | ISO | J:205449 | |||||||||
| Biological Process | GO:0043507 | positive regulation of JUN kinase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0043507 | positive regulation of JUN kinase activity | ISO | J:205449 | |||||||||
| Biological Process | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway | ISO | J:205449 | |||||||||
| Biological Process | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | ISO | J:205449 | |||||||||
| Biological Process | GO:1900745 | positive regulation of p38MAPK cascade | ISO | J:205449 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | ISO | J:205449 | |||||||||
| Biological Process | GO:0010632 | regulation of epithelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:1902498 | regulation of protein autoubiquitination | ISO | J:205449 | |||||||||
| Biological Process | GO:1900044 | regulation of protein K63-linked ubiquitination | ISO | J:205449 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||