|
Symbol Name ID |
Nudt12
nudix hydrolase 12 MGI:1915243 |
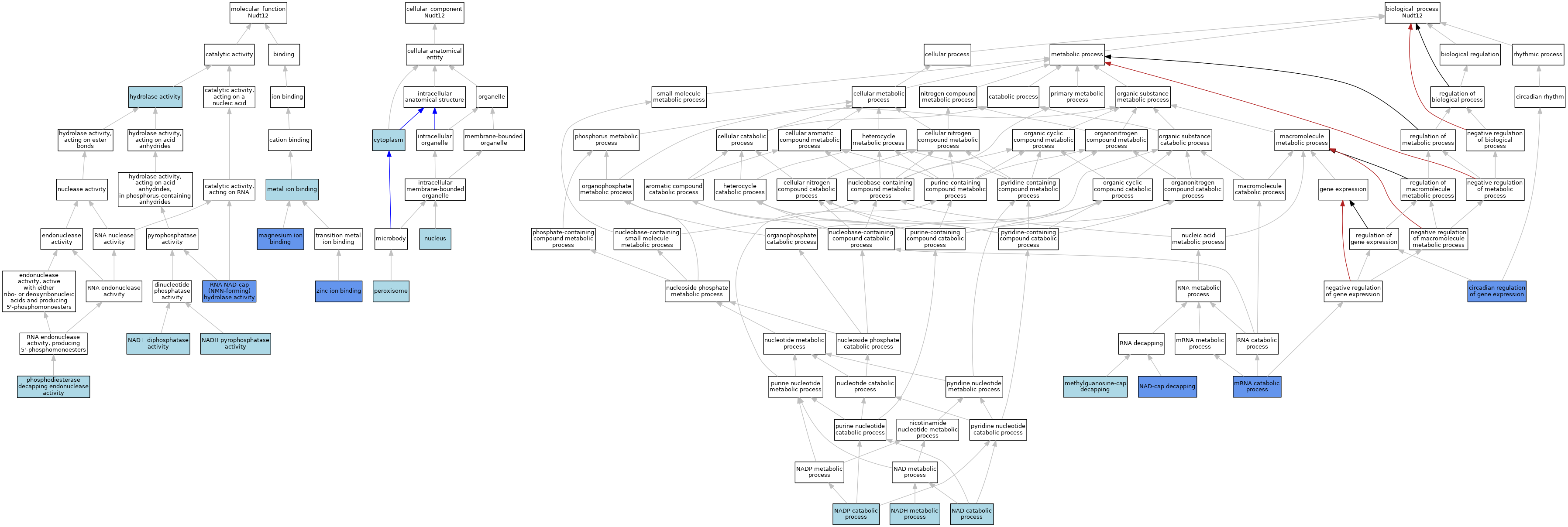
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000287 | magnesium ion binding | IDA | J:280091 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000210 | NAD+ diphosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000210 | NAD+ diphosphatase activity | ISO | J:178281 | |||||||||
| Molecular Function | GO:0035529 | NADH pyrophosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035529 | NADH pyrophosphatase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0035529 | NADH pyrophosphatase activity | ISO | J:178281 | |||||||||
| Molecular Function | GO:1990174 | phosphodiesterase decapping endonuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0110153 | RNA NAD-cap (NMN-forming) hydrolase activity | IDA | J:280091 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IDA | J:280091 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:178281 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005777 | peroxisome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005777 | peroxisome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005777 | peroxisome | ISO | J:178281 | |||||||||
| Biological Process | GO:0032922 | circadian regulation of gene expression | IMP | J:282288 | |||||||||
| Biological Process | GO:0110156 | methylguanosine-cap decapping | ISO | J:164563 | |||||||||
| Biological Process | GO:0006402 | mRNA catabolic process | IDA | J:280091 | |||||||||
| Biological Process | GO:0019677 | NAD catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0019677 | NAD catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0019677 | NAD catabolic process | ISO | J:178281 | |||||||||
| Biological Process | GO:0110155 | NAD-cap decapping | IDA | J:306601 | |||||||||
| Biological Process | GO:0110155 | NAD-cap decapping | ISO | J:164563 | |||||||||
| Biological Process | GO:0110155 | NAD-cap decapping | IDA | J:280091 | |||||||||
| Biological Process | GO:0006734 | NADH metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006742 | NADP catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006742 | NADP catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006742 | NADP catabolic process | ISO | J:178281 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||