|
Symbol Name ID |
Rab32
RAB32, member RAS oncogene family MGI:1915094 |
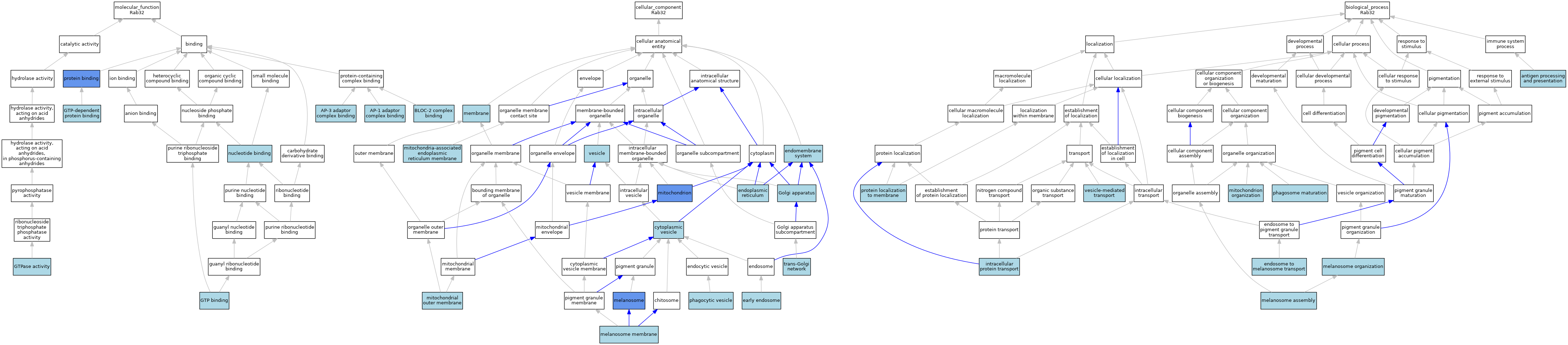
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0035650 | AP-1 adaptor complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035651 | AP-3 adaptor complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0036461 | BLOC-2 complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0030742 | GTP-dependent protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:215882 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005769 | early endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0012505 | endomembrane system | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IEA | J:72247 | |||||||||
| Cellular Component | GO:0042470 | melanosome | IDA | J:230050 | |||||||||
| Cellular Component | GO:0042470 | melanosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042470 | melanosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0033162 | melanosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0044233 | mitochondria-associated endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005741 | mitochondrial outer membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0045335 | phagocytic vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005802 | trans-Golgi network | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031982 | vesicle | IEA | J:72247 | |||||||||
| Biological Process | GO:0019882 | antigen processing and presentation | ISO | J:164563 | |||||||||
| Biological Process | GO:0035646 | endosome to melanosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006886 | intracellular protein transport | IBA | J:265628 | |||||||||
| Biological Process | GO:1903232 | melanosome assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0032438 | melanosome organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0007005 | mitochondrion organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0090382 | phagosome maturation | ISO | J:164563 | |||||||||
| Biological Process | GO:0072657 | protein localization to membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0016192 | vesicle-mediated transport | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||