|
Symbol Name ID |
Atg3
autophagy related 3 MGI:1915091 |
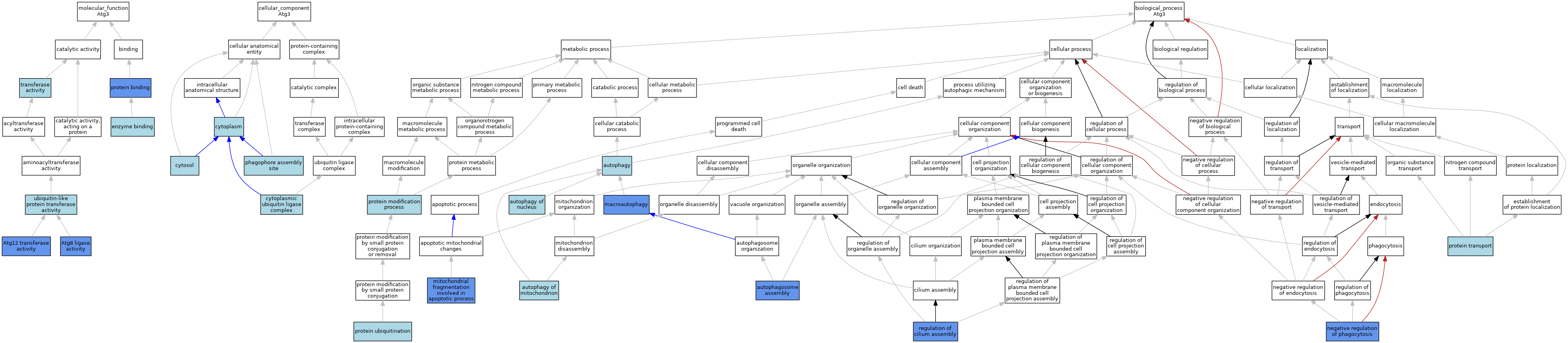
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019777 | Atg12 transferase activity | IDA | J:164739 | |||||||||
| Molecular Function | GO:0019776 | Atg8 ligase activity | IMP | J:164739 | |||||||||
| Molecular Function | GO:0019776 | Atg8 ligase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245241 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:164739 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0019787 | ubiquitin-like protein transferase activity | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000153 | cytoplasmic ubiquitin ligase complex | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000407 | phagophore assembly site | IBA | J:265628 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IMP | J:164739 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IMP | J:167845 | |||||||||
| Biological Process | GO:0006914 | autophagy | IEA | J:60000 | |||||||||
| Biological Process | GO:0000422 | autophagy of mitochondrion | IBA | J:265628 | |||||||||
| Biological Process | GO:0044804 | autophagy of nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0016236 | macroautophagy | IMP | J:208209 | |||||||||
| Biological Process | GO:0043653 | mitochondrial fragmentation involved in apoptotic process | IMP | J:164739 | |||||||||
| Biological Process | GO:0050765 | negative regulation of phagocytosis | IMP | J:208209 | |||||||||
| Biological Process | GO:0036211 | protein modification process | ISO | J:73065 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:1902017 | regulation of cilium assembly | IMP | J:203424 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||