|
Symbol Name ID |
Asph
aspartate-beta-hydroxylase MGI:1914186 |
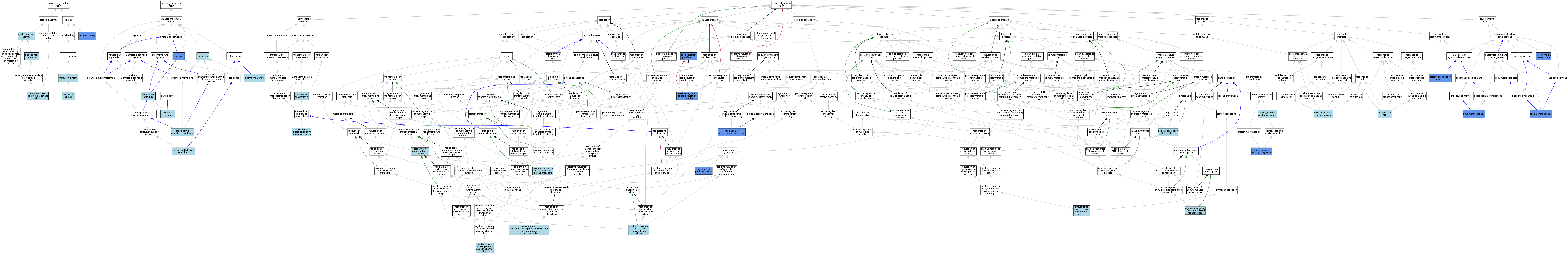
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051213 | dioxygenase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0062101 | peptidyl-aspartic acid 3-dioxygenase activity | ISS | J:66238 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:185106 | |||||||||
| Cellular Component | GO:0032541 | cortical endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032541 | cortical endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:185106 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISS | J:66238 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016529 | sarcoplasmic reticulum | IEA | J:60000 | |||||||||
| Biological Process | GO:0097202 | activation of cysteine-type endopeptidase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0032237 | activation of store-operated calcium channel activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0055074 | calcium ion homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0070588 | calcium ion transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IGI | J:75888 | |||||||||
| Biological Process | GO:0071277 | cellular response to calcium ion | ISO | J:164563 | |||||||||
| Biological Process | GO:0060325 | face morphogenesis | IMP | J:75888 | |||||||||
| Biological Process | GO:0035108 | limb morphogenesis | IMP | J:75888 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | IGI | J:75888 | |||||||||
| Biological Process | GO:0007389 | pattern specification process | IMP | J:75888 | |||||||||
| Biological Process | GO:0007389 | pattern specification process | IMP | J:75888 | |||||||||
| Biological Process | GO:0018193 | peptidyl-amino acid modification | IEA | J:72247 | |||||||||
| Biological Process | GO:0042264 | peptidyl-aspartic acid hydroxylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0042264 | peptidyl-aspartic acid hydroxylation | IDA | J:75888 | |||||||||
| Biological Process | GO:0010524 | positive regulation of calcium ion transport into cytosol | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0090316 | positive regulation of intracellular protein transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0045862 | positive regulation of proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0051480 | regulation of cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | ISO | J:164563 | |||||||||
| Biological Process | GO:1901879 | regulation of protein depolymerization | IDA | J:185106 | |||||||||
| Biological Process | GO:0031647 | regulation of protein stability | IDA | J:185106 | |||||||||
| Biological Process | GO:0033198 | response to ATP | ISO | J:164563 | |||||||||
| Biological Process | GO:0060021 | roof of mouth development | IMP | J:75888 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||