|
Symbol Name ID |
Dnajc10
DnaJ heat shock protein family (Hsp40) member C10 MGI:1914111 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
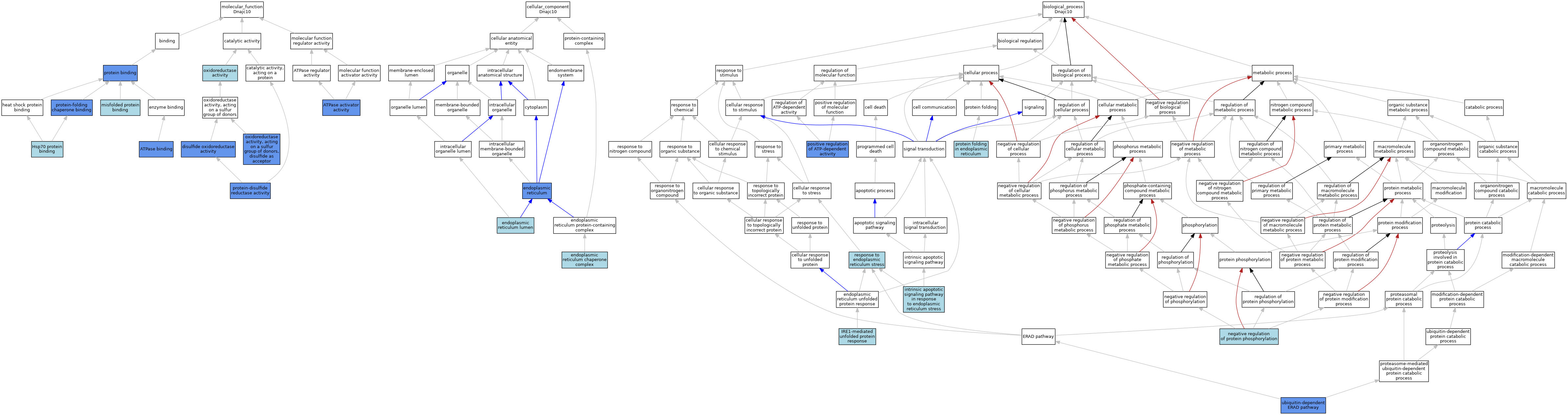
| Molecular Function | GO:0001671 | ATPase activator activity | IMP | J:81723 | |||||||||
| Molecular Function | GO:0051117 | ATPase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051117 | ATPase binding | IPI | J:81723 | |||||||||
| Molecular Function | GO:0015036 | disulfide oxidoreductase activity | IDA | J:169878 | |||||||||
| Molecular Function | GO:0030544 | Hsp70 protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051787 | misfolded protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051787 | misfolded protein binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor | IDA | J:170851 | |||||||||
| Molecular Function | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:170851 | |||||||||
| Molecular Function | GO:0015035 | protein-disulfide reductase activity | IDA | J:170851 | |||||||||
| Molecular Function | GO:0015035 | protein-disulfide reductase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | IDA | J:169878 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:81723 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:81369 | |||||||||
| Cellular Component | GO:0034663 | endoplasmic reticulum chaperone complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005788 | endoplasmic reticulum lumen | ISO | J:164563 | |||||||||
| Biological Process | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | ISO | J:164563 | |||||||||
| Biological Process | GO:0036498 | IRE1-mediated unfolded protein response | IBA | J:265628 | |||||||||
| Biological Process | GO:0001933 | negative regulation of protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0032781 | positive regulation of ATP-dependent activity | IMP | J:81723 | |||||||||
| Biological Process | GO:0034975 | protein folding in endoplasmic reticulum | ISO | J:164563 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | ISO | J:164563 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | IDA | J:170851 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | IDA | J:169878 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||