|
Symbol Name ID |
Retreg1
reticulophagy regulator 1 MGI:1913520 |
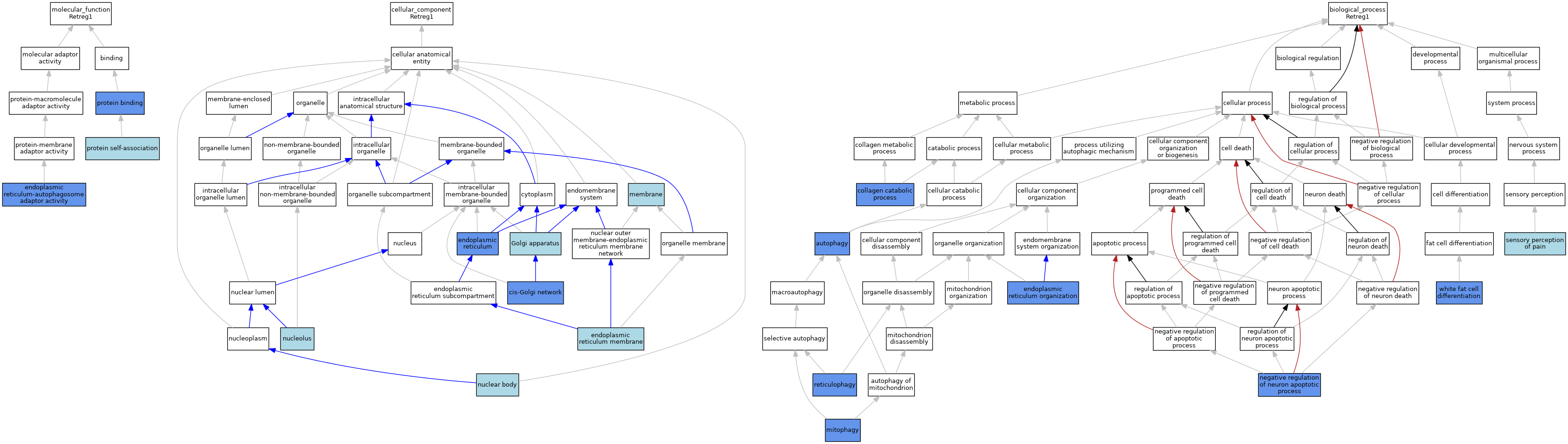
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0140506 | endoplasmic reticulum-autophagosome adaptor activity | IMP | J:313627 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:313627 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:313627 | |||||||||
| Molecular Function | GO:0043621 | protein self-association | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005801 | cis-Golgi network | IDA | J:154862 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:313627 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016604 | nuclear body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006914 | autophagy | IDA | J:280916 | |||||||||
| Biological Process | GO:0030574 | collagen catabolic process | IMP | J:313627 | |||||||||
| Biological Process | GO:0007029 | endoplasmic reticulum organization | IMP | J:313627 | |||||||||
| Biological Process | GO:0000423 | mitophagy | IDA | J:280916 | |||||||||
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | IMP | J:222922 | |||||||||
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | IMP | J:222922 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | IBA | J:265628 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | IMP | J:313627 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | IDA | J:313627 | |||||||||
| Biological Process | GO:0019233 | sensory perception of pain | ISO | J:164563 | |||||||||
| Biological Process | GO:0050872 | white fat cell differentiation | IDA | J:280916 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||