|
Symbol Name ID |
Wasf1
WASP family, member 1 MGI:1890563 |
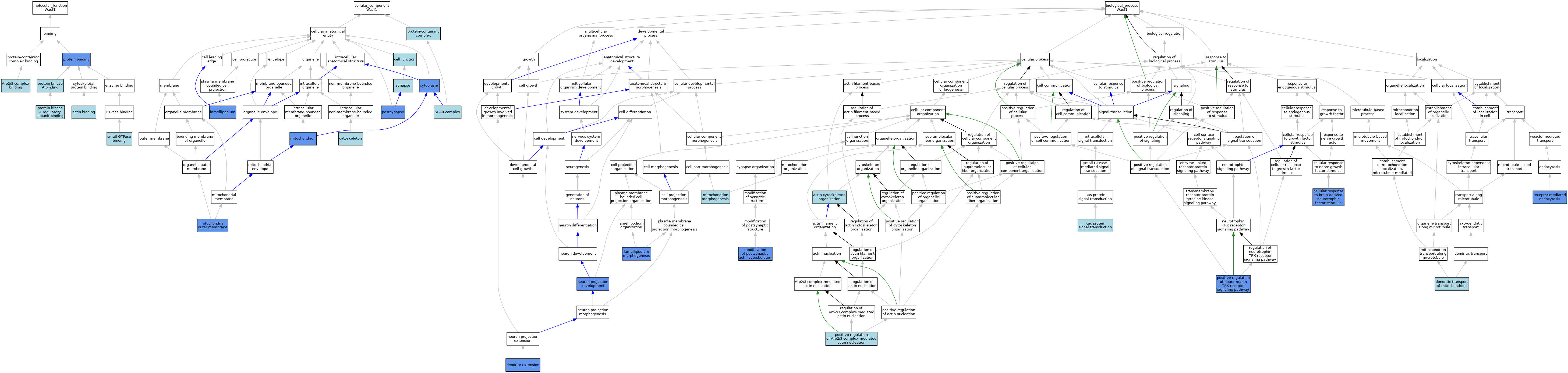
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003779 | actin binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0071933 | Arp2/3 complex binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:224720 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:180006 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:206834 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:85083 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:85083 | |||||||||
| Molecular Function | GO:0051018 | protein kinase A binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0034237 | protein kinase A regulatory subunit binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:149119 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IDA | J:91137 | |||||||||
| Cellular Component | GO:0005741 | mitochondrial outer membrane | IDA | J:85083 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:85083 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:85083 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IDA | J:263433 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IMP | J:263433 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IDA | J:263433 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031209 | SCAR complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031209 | SCAR complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0045202 | synapse | IEA | J:60000 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | IBA | J:265628 | |||||||||
| Biological Process | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | IMP | J:239183 | |||||||||
| Biological Process | GO:0097484 | dendrite extension | IMP | J:239183 | |||||||||
| Biological Process | GO:0098939 | dendritic transport of mitochondrion | ISO | J:155856 | |||||||||
| Biological Process | GO:0072673 | lamellipodium morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0072673 | lamellipodium morphogenesis | IMP | J:149119 | |||||||||
| Biological Process | GO:0070584 | mitochondrion morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IPI | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IDA | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IPI | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IMP | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IDA | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IDA | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IDA | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IMP | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IMP | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IMP | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IMP | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IDA | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IMP | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IPI | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IDA | J:263395 | |||||||||
| Biological Process | GO:0098885 | modification of postsynaptic actin cytoskeleton | IPI | J:263395 | |||||||||
| Biological Process | GO:0031175 | neuron projection development | IMP | J:239183 | |||||||||
| Biological Process | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | ISO | J:164563 | |||||||||
| Biological Process | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | IBA | J:265628 | |||||||||
| Biological Process | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway | IMP | J:239183 | |||||||||
| Biological Process | GO:0016601 | Rac protein signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0006898 | receptor-mediated endocytosis | IMP | J:239183 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||