|
Symbol Name ID |
Larp1
La ribonucleoprotein 1, translational regulator MGI:1890165 |
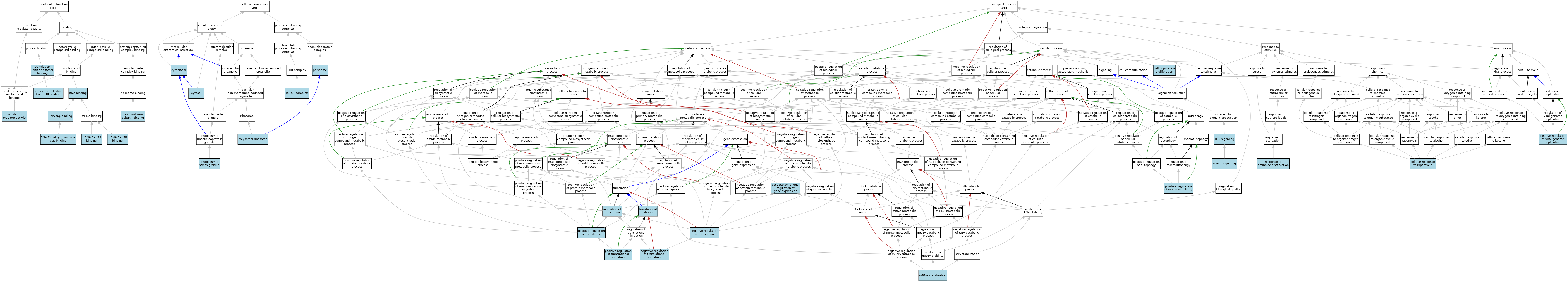
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008190 | eukaryotic initiation factor 4E binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003730 | mRNA 3'-UTR binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0048027 | mRNA 5'-UTR binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043024 | ribosomal small subunit binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000340 | RNA 7-methylguanosine cap binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000339 | RNA cap binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008494 | translation activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031369 | translation initiation factor binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISA | J:63224 | |||||||||
| Cellular Component | GO:0010494 | cytoplasmic stress granule | ISO | J:164563 | |||||||||
| Cellular Component | GO:0010494 | cytoplasmic stress granule | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0042788 | polysomal ribosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005844 | polysome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031931 | TORC1 complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0072752 | cellular response to rapamycin | ISO | J:164563 | |||||||||
| Biological Process | GO:0048255 | mRNA stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0017148 | negative regulation of translation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045947 | negative regulation of translational initiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0016239 | positive regulation of macroautophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0045727 | positive regulation of translation | IBA | J:265628 | |||||||||
| Biological Process | GO:0045948 | positive regulation of translational initiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045070 | positive regulation of viral genome replication | ISO | J:164563 | |||||||||
| Biological Process | GO:0010608 | post-transcriptional regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:0006417 | regulation of translation | IEA | J:60000 | |||||||||
| Biological Process | GO:1990928 | response to amino acid starvation | ISO | J:164563 | |||||||||
| Biological Process | GO:0031929 | TOR signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0038202 | TORC1 signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0006413 | translational initiation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||