|
Symbol Name ID |
Ttyh1
tweety family member 1 MGI:1889007 |
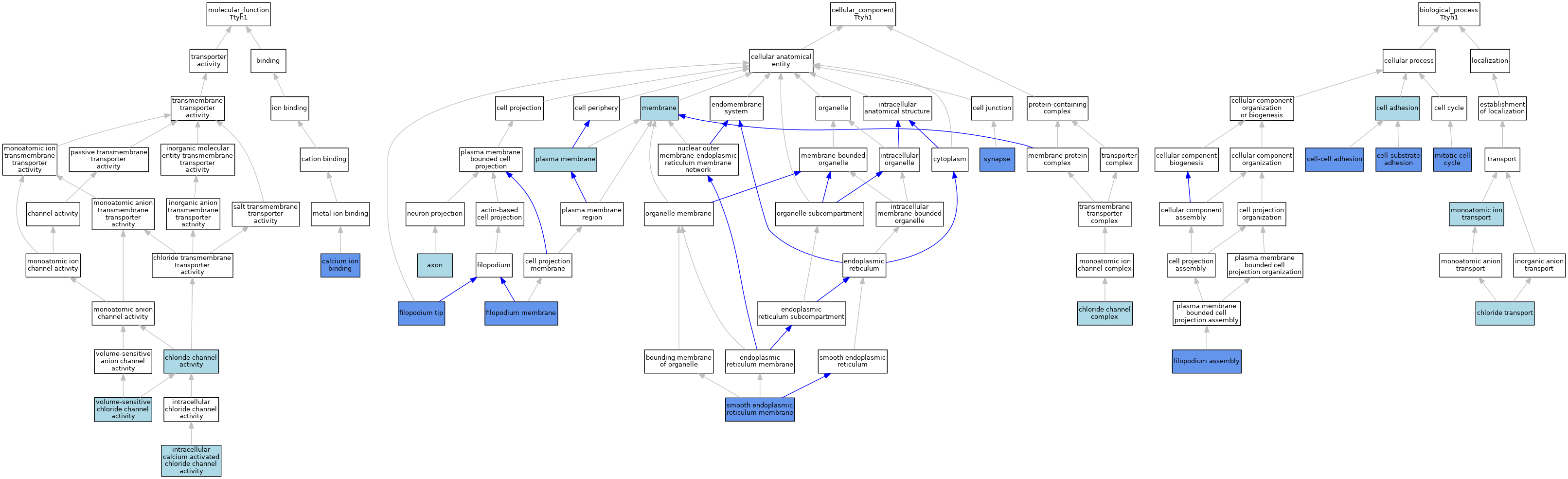
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | IDA | J:161769 | |||||||||
| Molecular Function | GO:0005254 | chloride channel activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005229 | intracellular calcium activated chloride channel activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0072320 | volume-sensitive chloride channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0072320 | volume-sensitive chloride channel activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030424 | axon | ISO | J:73065 | |||||||||
| Cellular Component | GO:0030424 | axon | ISO | J:73065 | |||||||||
| Cellular Component | GO:0034707 | chloride channel complex | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031527 | filopodium membrane | IDA | J:119553 | |||||||||
| Cellular Component | GO:0032433 | filopodium tip | IDA | J:119553 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030868 | smooth endoplasmic reticulum membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030868 | smooth endoplasmic reticulum membrane | IDA | J:161796 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:182391 | |||||||||
| Cellular Component | GO:0045202 | synapse | EXP | J:182391 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IEA | J:60000 | |||||||||
| Biological Process | GO:0098609 | cell-cell adhesion | IDA | J:119553 | |||||||||
| Biological Process | GO:0031589 | cell-substrate adhesion | IDA | J:119553 | |||||||||
| Biological Process | GO:0006821 | chloride transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0046847 | filopodium assembly | IDA | J:119553 | |||||||||
| Biological Process | GO:0000278 | mitotic cell cycle | IMP | J:161796 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||