|
Symbol Name ID |
Fut8
fucosyltransferase 8 MGI:1858901 |
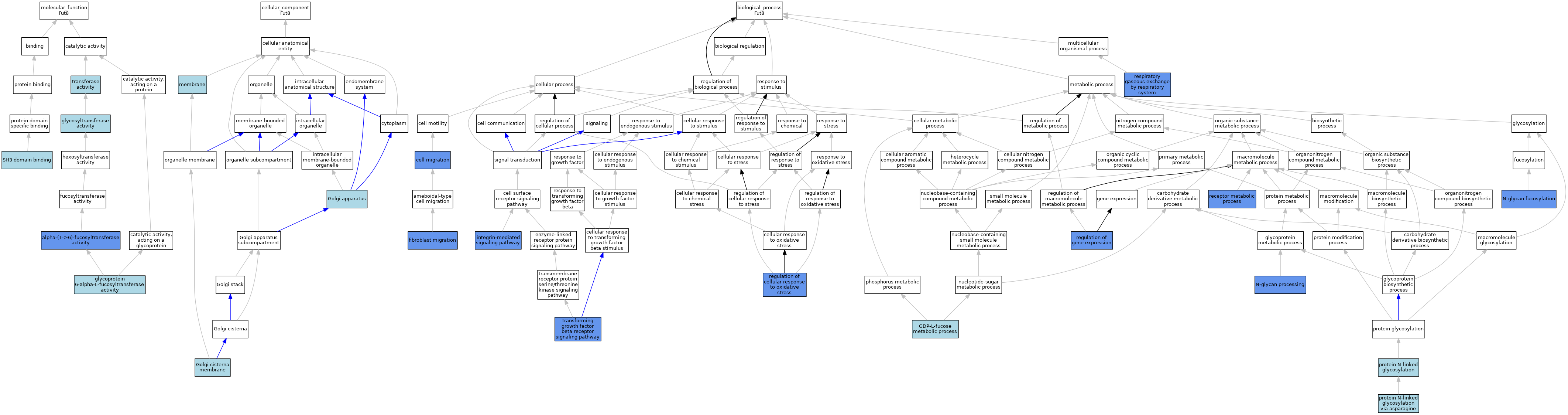
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0046921 | alpha-(1->6)-fucosyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0046921 | alpha-(1->6)-fucosyltransferase activity | IMP | J:102932 | |||||||||
| Molecular Function | GO:0046921 | alpha-(1->6)-fucosyltransferase activity | IMP | J:102932 | |||||||||
| Molecular Function | GO:0046921 | alpha-(1->6)-fucosyltransferase activity | IMP | J:102932 | |||||||||
| Molecular Function | GO:0046921 | alpha-(1->6)-fucosyltransferase activity | IMP | J:117643 | |||||||||
| Molecular Function | GO:0046921 | alpha-(1->6)-fucosyltransferase activity | IMP | J:117643 | |||||||||
| Molecular Function | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016757 | glycosyltransferase activity | ISA | J:74082 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0032580 | Golgi cisterna membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Biological Process | GO:0016477 | cell migration | IMP | J:117643 | |||||||||
| Biological Process | GO:0010761 | fibroblast migration | IMP | J:117643 | |||||||||
| Biological Process | GO:0046368 | GDP-L-fucose metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0007229 | integrin-mediated signaling pathway | IMP | J:117643 | |||||||||
| Biological Process | GO:0007229 | integrin-mediated signaling pathway | IMP | J:117643 | |||||||||
| Biological Process | GO:0036071 | N-glycan fucosylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0036071 | N-glycan fucosylation | IMP | J:217242 | |||||||||
| Biological Process | GO:0006491 | N-glycan processing | IMP | J:217242 | |||||||||
| Biological Process | GO:0006487 | protein N-linked glycosylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0018279 | protein N-linked glycosylation via asparagine | ISO | J:164563 | |||||||||
| Biological Process | GO:0043112 | receptor metabolic process | IMP | J:102932 | |||||||||
| Biological Process | GO:1900407 | regulation of cellular response to oxidative stress | IMP | J:217242 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IMP | J:217242 | |||||||||
| Biological Process | GO:0007585 | respiratory gaseous exchange by respiratory system | IMP | J:102932 | |||||||||
| Biological Process | GO:0007179 | transforming growth factor beta receptor signaling pathway | IMP | J:102932 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||