|
Symbol Name ID |
Nckap1
NCK-associated protein 1 MGI:1355333 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
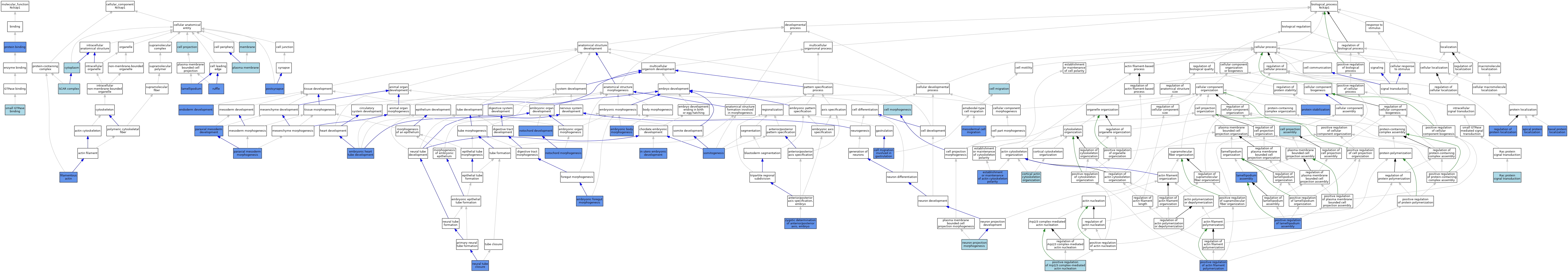
| Molecular Function | GO:0005515 | protein binding | IPI | J:180006 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:180006 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031941 | filamentous actin | IDA | J:241033 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IDA | J:241033 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IDA | J:88449 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IEP | J:201819 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IDA | J:201819 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IDA | J:201819 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IDA | J:201819 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | IDA | J:201819 | |||||||||
| Cellular Component | GO:0001726 | ruffle | IDA | J:241033 | |||||||||
| Cellular Component | GO:0031209 | SCAR complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031209 | SCAR complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0045176 | apical protein localization | IDA | J:119028 | |||||||||
| Biological Process | GO:0045176 | apical protein localization | IDA | J:119028 | |||||||||
| Biological Process | GO:0045176 | apical protein localization | IDA | J:119028 | |||||||||
| Biological Process | GO:0045175 | basal protein localization | IDA | J:119028 | |||||||||
| Biological Process | GO:0045175 | basal protein localization | IDA | J:119028 | |||||||||
| Biological Process | GO:0045175 | basal protein localization | IDA | J:119028 | |||||||||
| Biological Process | GO:0016477 | cell migration | IBA | J:265628 | |||||||||
| Biological Process | GO:0042074 | cell migration involved in gastrulation | IMP | J:119028 | |||||||||
| Biological Process | GO:0042074 | cell migration involved in gastrulation | IMP | J:119028 | |||||||||
| Biological Process | GO:0000902 | cell morphogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0030031 | cell projection assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0030866 | cortical actin cytoskeleton organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0010172 | embryonic body morphogenesis | IMP | J:119028 | |||||||||
| Biological Process | GO:0048617 | embryonic foregut morphogenesis | IMP | J:119028 | |||||||||
| Biological Process | GO:0035050 | embryonic heart tube development | IMP | J:119028 | |||||||||
| Biological Process | GO:0007492 | endoderm development | IMP | J:119028 | |||||||||
| Biological Process | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity | IMP | J:119028 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IMP | J:119028 | |||||||||
| Biological Process | GO:0030032 | lamellipodium assembly | IDA | J:88449 | |||||||||
| Biological Process | GO:0030032 | lamellipodium assembly | IMP | J:119028 | |||||||||
| Biological Process | GO:0008078 | mesodermal cell migration | IMP | J:119028 | |||||||||
| Biological Process | GO:0001843 | neural tube closure | IMP | J:119028 | |||||||||
| Biological Process | GO:0048812 | neuron projection morphogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0030903 | notochord development | IMP | J:119028 | |||||||||
| Biological Process | GO:0048570 | notochord morphogenesis | IMP | J:119028 | |||||||||
| Biological Process | GO:0048339 | paraxial mesoderm development | IMP | J:119028 | |||||||||
| Biological Process | GO:0048339 | paraxial mesoderm development | IMP | J:119028 | |||||||||
| Biological Process | GO:0048340 | paraxial mesoderm morphogenesis | IMP | J:119028 | |||||||||
| Biological Process | GO:0030838 | positive regulation of actin filament polymerization | IDA | J:241033 | |||||||||
| Biological Process | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | ISO | J:164563 | |||||||||
| Biological Process | GO:0010592 | positive regulation of lamellipodium assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0010592 | positive regulation of lamellipodium assembly | IDA | J:241033 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | IMP | J:119028 | |||||||||
| Biological Process | GO:0016601 | Rac protein signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0032880 | regulation of protein localization | IMP | J:119028 | |||||||||
| Biological Process | GO:0001756 | somitogenesis | IMP | J:119028 | |||||||||
| Biological Process | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | IMP | J:119028 | |||||||||
| Biological Process | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | IMP | J:119028 | |||||||||
| Biological Process | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | IMP | J:119028 | |||||||||
| Biological Process | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | IMP | J:119028 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||