|
Symbol Name ID |
Il27ra
interleukin 27 receptor, alpha MGI:1355318 |
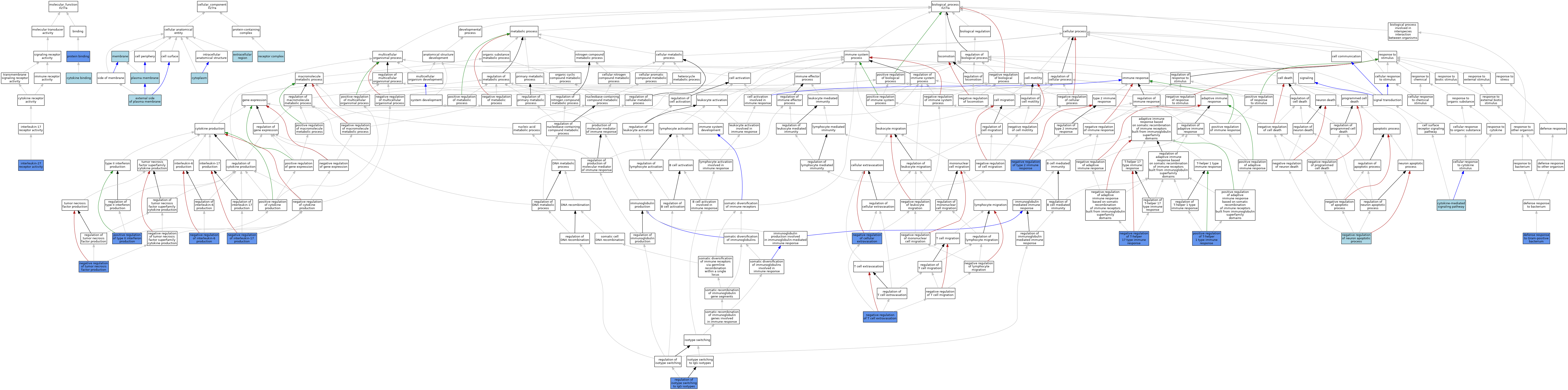
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019955 | cytokine binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0045509 | interleukin-27 receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0045509 | interleukin-27 receptor activity | IDA | J:77522 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:77522 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:77522 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | TAS | Reactome:R-MMU-8950729 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | TAS | Reactome:R-MMU-8950729 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-8950409 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IMP | J:65439 | |||||||||
| Biological Process | GO:0002692 | negative regulation of cellular extravasation | IGI | J:212625 | |||||||||
| Biological Process | GO:0032700 | negative regulation of interleukin-17 production | IGI | J:212625 | |||||||||
| Biological Process | GO:0032715 | negative regulation of interleukin-6 production | IGI | J:212625 | |||||||||
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:2000408 | negative regulation of T cell extravasation | IGI | J:212625 | |||||||||
| Biological Process | GO:2000317 | negative regulation of T-helper 17 type immune response | IGI | J:212625 | |||||||||
| Biological Process | GO:0032720 | negative regulation of tumor necrosis factor production | IGI | J:212625 | |||||||||
| Biological Process | GO:0002829 | negative regulation of type 2 immune response | IMP | J:65439 | |||||||||
| Biological Process | GO:0002827 | positive regulation of T-helper 1 type immune response | IMP | J:65439 | |||||||||
| Biological Process | GO:0032729 | positive regulation of type II interferon production | IMP | J:65439 | |||||||||
| Biological Process | GO:0048302 | regulation of isotype switching to IgG isotypes | IMP | J:65439 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||