|
Symbol Name ID |
Fkbp8
FK506 binding protein 8 MGI:1341070 |
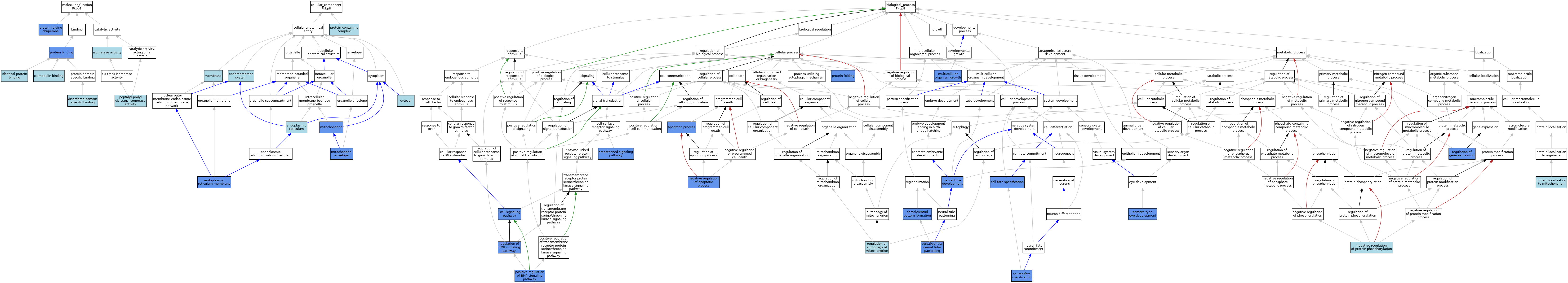
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005516 | calmodulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0097718 | disordered domain specific binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016853 | isomerase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:272245 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:272245 | |||||||||
| Molecular Function | GO:0044183 | protein folding chaperone | IBA | J:265628 | |||||||||
| Molecular Function | GO:0044183 | protein folding chaperone | IMP | J:272245 | |||||||||
| Molecular Function | GO:0044183 | protein folding chaperone | ISO | J:272245 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0012505 | endomembrane system | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IDA | J:89364 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005740 | mitochondrial envelope | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005740 | mitochondrial envelope | IDA | J:89364 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IMP | J:132031 | |||||||||
| Biological Process | GO:0030509 | BMP signaling pathway | IMP | J:138713 | |||||||||
| Biological Process | GO:0043010 | camera-type eye development | IMP | J:89364 | |||||||||
| Biological Process | GO:0001708 | cell fate specification | IMP | J:138713 | |||||||||
| Biological Process | GO:0021904 | dorsal/ventral neural tube patterning | IGI | J:138713 | |||||||||
| Biological Process | GO:0021904 | dorsal/ventral neural tube patterning | IMP | J:138713 | |||||||||
| Biological Process | GO:0021904 | dorsal/ventral neural tube patterning | IGI | J:138713 | |||||||||
| Biological Process | GO:0021904 | dorsal/ventral neural tube patterning | IMP | J:132031 | |||||||||
| Biological Process | GO:0021904 | dorsal/ventral neural tube patterning | IDA | J:138713 | |||||||||
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | IMP | J:89364 | |||||||||
| Biological Process | GO:0035264 | multicellular organism growth | IGI | J:138713 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IMP | J:132031 | |||||||||
| Biological Process | GO:0001933 | negative regulation of protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0021915 | neural tube development | IMP | J:138713 | |||||||||
| Biological Process | GO:0048665 | neuron fate specification | IMP | J:89364 | |||||||||
| Biological Process | GO:0030513 | positive regulation of BMP signaling pathway | IMP | J:138713 | |||||||||
| Biological Process | GO:0006457 | protein folding | IBA | J:265628 | |||||||||
| Biological Process | GO:0006457 | protein folding | ISO | J:272245 | |||||||||
| Biological Process | GO:0006457 | protein folding | IMP | J:272245 | |||||||||
| Biological Process | GO:0070585 | protein localization to mitochondrion | ISO | J:164563 | |||||||||
| Biological Process | GO:1903146 | regulation of autophagy of mitochondrion | ISO | J:164563 | |||||||||
| Biological Process | GO:0030510 | regulation of BMP signaling pathway | IGI | J:138713 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IGI | J:138713 | |||||||||
| Biological Process | GO:0007224 | smoothened signaling pathway | IMP | J:89364 | |||||||||
| Biological Process | GO:0007224 | smoothened signaling pathway | IGI | J:89364 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||