|
Symbol Name ID |
Kcnq2
potassium voltage-gated channel, subfamily Q, member 2 MGI:1309503 |
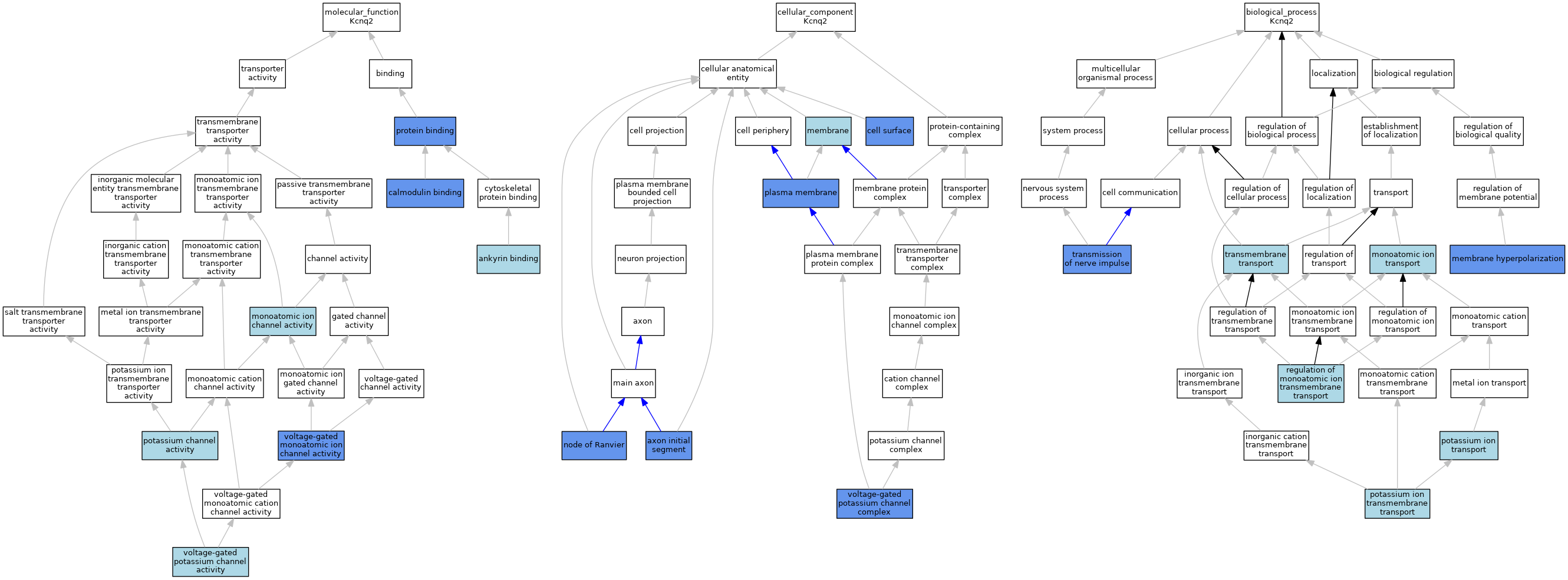
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0030506 | ankyrin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005516 | calmodulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005516 | calmodulin binding | IPI | J:79145 | |||||||||
| Molecular Function | GO:0005216 | monoatomic ion channel activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005267 | potassium channel activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:240720 | |||||||||
| Molecular Function | GO:0005244 | voltage-gated monoatomic ion channel activity | IGI | J:79145 | |||||||||
| Molecular Function | GO:0005249 | voltage-gated potassium channel activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005249 | voltage-gated potassium channel activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005249 | voltage-gated potassium channel activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043194 | axon initial segment | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043194 | axon initial segment | IDA | J:106271 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:79145 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0033268 | node of Ranvier | IDA | J:106271 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:106271 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0008076 | voltage-gated potassium channel complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0008076 | voltage-gated potassium channel complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0008076 | voltage-gated potassium channel complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0008076 | voltage-gated potassium channel complex | IGI | J:79145 | |||||||||
| Biological Process | GO:0060081 | membrane hyperpolarization | IMP | MGI:MGI:62459 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0006813 | potassium ion transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0006813 | potassium ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0034765 | regulation of monoatomic ion transmembrane transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0055085 | transmembrane transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0019226 | transmission of nerve impulse | IMP | J:62797 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||