|
Symbol Name ID |
Bscl2
BSCL2 lipid droplet biogenesis associated, seipin MGI:1298392 |
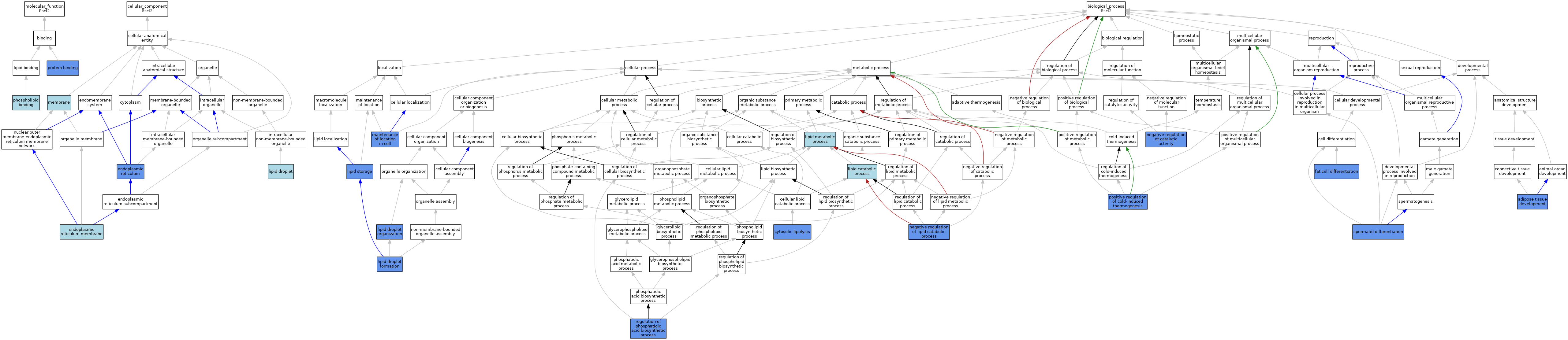
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005543 | phospholipid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:242338 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:242338 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:139166 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:198139 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005811 | lipid droplet | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Biological Process | GO:0060612 | adipose tissue development | IMP | J:173404 | |||||||||
| Biological Process | GO:0061725 | cytosolic lipolysis | IMP | J:229489 | |||||||||
| Biological Process | GO:0045444 | fat cell differentiation | IMP | J:183701 | |||||||||
| Biological Process | GO:0045444 | fat cell differentiation | IMP | J:198139 | |||||||||
| Biological Process | GO:0016042 | lipid catabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0140042 | lipid droplet formation | ISO | J:164563 | |||||||||
| Biological Process | GO:0140042 | lipid droplet formation | IDA | J:198139 | |||||||||
| Biological Process | GO:0034389 | lipid droplet organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0034389 | lipid droplet organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0034389 | lipid droplet organization | IMP | J:242338 | |||||||||
| Biological Process | GO:0034389 | lipid droplet organization | IMP | J:211072 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0019915 | lipid storage | ISO | J:164563 | |||||||||
| Biological Process | GO:0019915 | lipid storage | IBA | J:265628 | |||||||||
| Biological Process | GO:0019915 | lipid storage | IMP | J:198139 | |||||||||
| Biological Process | GO:0051651 | maintenance of location in cell | IMP | J:198139 | |||||||||
| Biological Process | GO:0043086 | negative regulation of catalytic activity | IMP | J:242338 | |||||||||
| Biological Process | GO:0043086 | negative regulation of catalytic activity | IMP | J:242338 | |||||||||
| Biological Process | GO:0050995 | negative regulation of lipid catabolic process | IMP | J:183701 | |||||||||
| Biological Process | GO:0120162 | positive regulation of cold-induced thermogenesis | IMP | J:249911 | |||||||||
| Biological Process | GO:1905693 | regulation of phosphatidic acid biosynthetic process | IMP | J:242338 | |||||||||
| Biological Process | GO:0048515 | spermatid differentiation | IMP | J:211072 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||