|
Symbol Name ID |
Cldn1
claudin 1 MGI:1276109 |
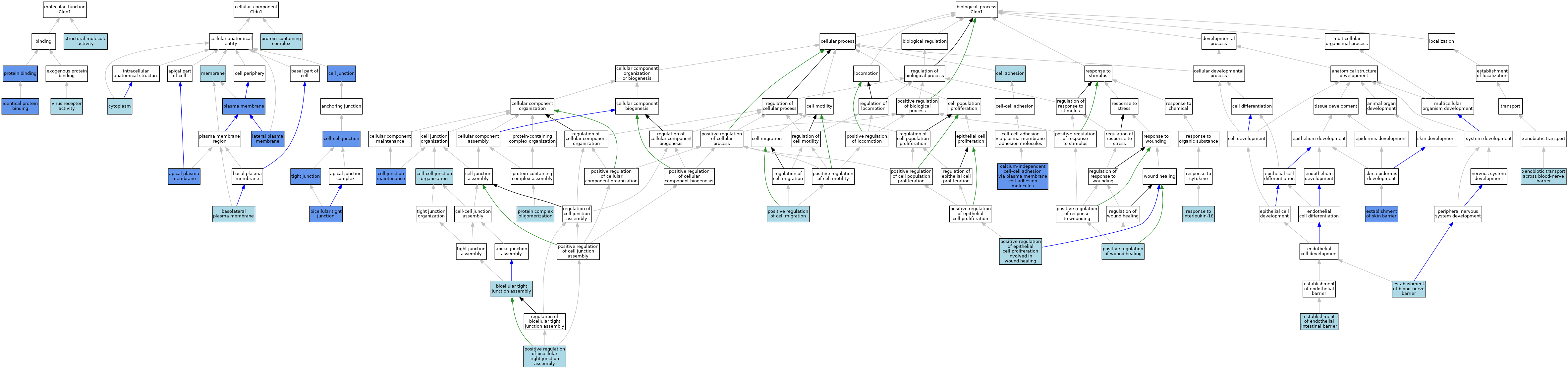
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:94758 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:58914 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94758 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:73641 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:73641 | |||||||||
| Molecular Function | GO:0005198 | structural molecule activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0001618 | virus receptor activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | IDA | J:159723 | |||||||||
| Cellular Component | GO:0016323 | basolateral plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:75447 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:48635 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:200284 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:104866 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:107229 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:181271 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:97223 | |||||||||
| Cellular Component | GO:0005923 | bicellular tight junction | IDA | J:181271 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IDA | J:219071 | |||||||||
| Cellular Component | GO:0030054 | cell junction | TAS | Reactome:R-MMU-420000 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | IDA | J:280103 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016328 | lateral plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016328 | lateral plasma membrane | IDA | J:181271 | |||||||||
| Cellular Component | GO:0016020 | membrane | NAS | J:48635 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:75447 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-420000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:97223 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0070160 | tight junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0070160 | tight junction | IDA | J:280385 | |||||||||
| Biological Process | GO:0070830 | bicellular tight junction assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0070830 | bicellular tight junction assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | IDA | J:113790 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0034331 | cell junction maintenance | IMP | J:280103 | |||||||||
| Biological Process | GO:0045216 | cell-cell junction organization | ISO | J:73065 | |||||||||
| Biological Process | GO:0008065 | establishment of blood-nerve barrier | ISO | J:155856 | |||||||||
| Biological Process | GO:0090557 | establishment of endothelial intestinal barrier | ISO | J:155856 | |||||||||
| Biological Process | GO:0061436 | establishment of skin barrier | IMP | J:75447 | |||||||||
| Biological Process | GO:0061436 | establishment of skin barrier | ISO | J:164563 | |||||||||
| Biological Process | GO:1903348 | positive regulation of bicellular tight junction assembly | ISO | J:155856 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing | ISO | J:164563 | |||||||||
| Biological Process | GO:0090303 | positive regulation of wound healing | ISO | J:164563 | |||||||||
| Biological Process | GO:0051259 | protein complex oligomerization | ISO | J:164563 | |||||||||
| Biological Process | GO:0070673 | response to interleukin-18 | ISO | J:155856 | |||||||||
| Biological Process | GO:0061772 | xenobiotic transport across blood-nerve barrier | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||