|
Symbol Name ID |
Hap1
huntingtin-associated protein 1 MGI:1261831 |
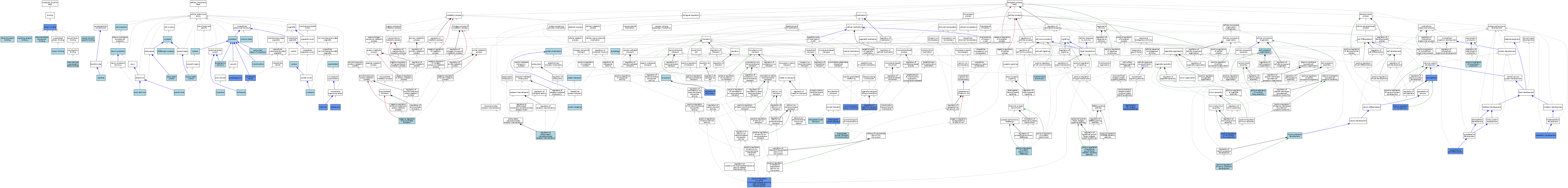
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0048403 | brain-derived neurotrophic factor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0017022 | myosin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:198594 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:175180 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:169046 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:91294 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:91294 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:47245 | |||||||||
| Molecular Function | GO:0019904 | protein domain specific binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0044325 | transmembrane transporter binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005776 | autophagosome | IDA | J:206857 | |||||||||
| Cellular Component | GO:0043679 | axon terminus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005814 | centriole | IDA | J:178446 | |||||||||
| Cellular Component | GO:0005813 | centrosome | IDA | J:178446 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IDA | J:91294 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030425 | dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0098982 | GABA-ergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030426 | growth cone | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016234 | inclusion body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0099524 | postsynaptic cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0099523 | presynaptic cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IEA | J:60000 | |||||||||
| Biological Process | GO:0008089 | anterograde axonal transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0008089 | anterograde axonal transport | IMP | J:208015 | |||||||||
| Biological Process | GO:0098957 | anterograde axonal transport of mitochondrion | IBA | J:265628 | |||||||||
| Biological Process | GO:0006914 | autophagy | IEA | J:60000 | |||||||||
| Biological Process | GO:0030030 | cell projection organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0021549 | cerebellum development | IMP | J:140869 | |||||||||
| Biological Process | GO:0006887 | exocytosis | IEA | J:60000 | |||||||||
| Biological Process | GO:0021979 | hypothalamus cell differentiation | IMP | J:208015 | |||||||||
| Biological Process | GO:0048311 | mitochondrion distribution | IBA | J:265628 | |||||||||
| Biological Process | GO:1902430 | negative regulation of amyloid-beta formation | ISO | J:155856 | |||||||||
| Biological Process | GO:0022008 | neurogenesis | IMP | J:140869 | |||||||||
| Biological Process | GO:0022008 | neurogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0031175 | neuron projection development | ISO | J:155856 | |||||||||
| Biological Process | GO:0048011 | neurotrophin TRK receptor signaling pathway | IMP | J:208015 | |||||||||
| Biological Process | GO:0048011 | neurotrophin TRK receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0090261 | positive regulation of inclusion body assembly | ISO | J:155856 | |||||||||
| Biological Process | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | IMP | J:101292 | |||||||||
| Biological Process | GO:0050769 | positive regulation of neurogenesis | IMP | J:208015 | |||||||||
| Biological Process | GO:0010976 | positive regulation of neuron projection development | ISO | J:155856 | |||||||||
| Biological Process | GO:0032901 | positive regulation of neurotrophin production | ISO | J:155856 | |||||||||
| Biological Process | GO:1902857 | positive regulation of non-motile cilium assembly | IMP | J:178446 | |||||||||
| Biological Process | GO:0032230 | positive regulation of synaptic transmission, GABAergic | ISO | J:155856 | |||||||||
| Biological Process | GO:0008104 | protein localization | ISO | J:164563 | |||||||||
| Biological Process | GO:0006605 | protein targeting | IBA | J:265628 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0017157 | regulation of exocytosis | IMP | J:213185 | |||||||||
| Biological Process | GO:1902513 | regulation of organelle transport along microtubule | IMP | J:206857 | |||||||||
| Biological Process | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | ISO | J:155856 | |||||||||
| Biological Process | GO:0008090 | retrograde axonal transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0047496 | vesicle transport along microtubule | IBA | J:265628 | |||||||||
| Biological Process | GO:0047496 | vesicle transport along microtubule | IMP | J:91294 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||