|
Symbol Name ID |
Nrdc
nardilysin convertase MGI:1201386 |
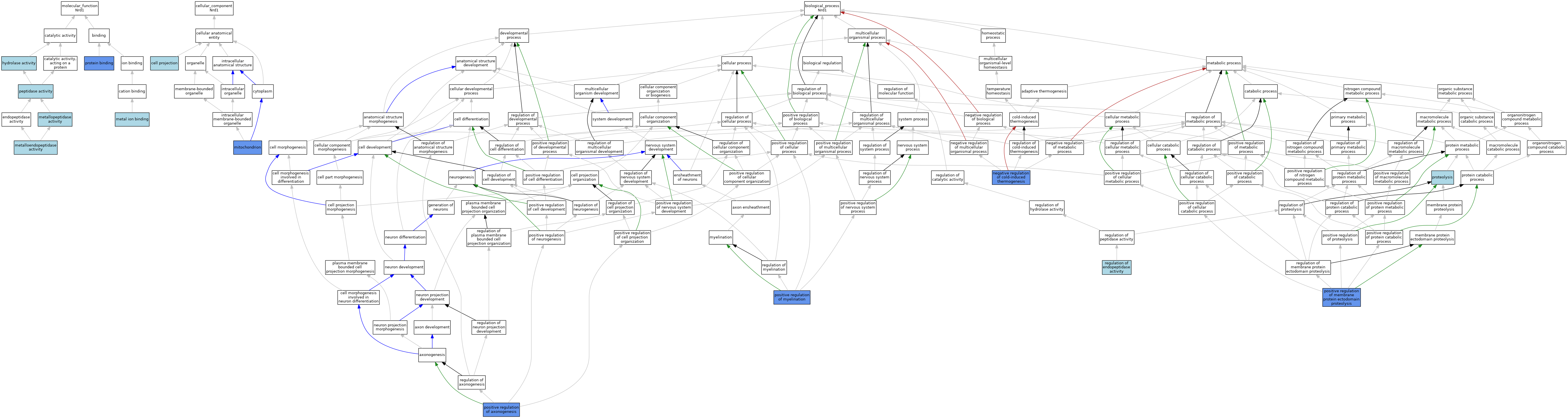
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0008237 | metallopeptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | ISO | J:100124 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:155532 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:100124 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Biological Process | GO:0120163 | negative regulation of cold-induced thermogenesis | IMP | J:225248 | |||||||||
| Biological Process | GO:0050772 | positive regulation of axonogenesis | IMP | J:155532 | |||||||||
| Biological Process | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | IDA | J:155532 | |||||||||
| Biological Process | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0031643 | positive regulation of myelination | IMP | J:155532 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:72247 | |||||||||
| Biological Process | GO:0052548 | regulation of endopeptidase activity | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||