|
Symbol Name ID |
Polg
polymerase (DNA directed), gamma MGI:1196389 |
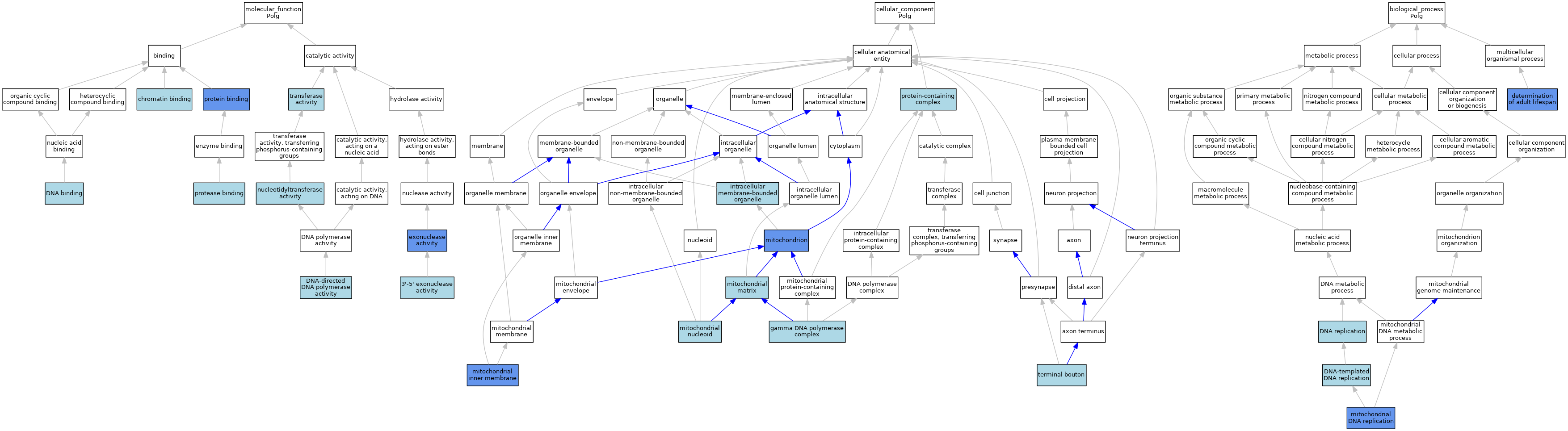
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008408 | 3'-5' exonuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008408 | 3'-5' exonuclease activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003887 | DNA-directed DNA polymerase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003887 | DNA-directed DNA polymerase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003887 | DNA-directed DNA polymerase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003887 | DNA-directed DNA polymerase activity | ISO | J:91077 | |||||||||
| Molecular Function | GO:0004527 | exonuclease activity | IDA | J:91077 | |||||||||
| Molecular Function | GO:0016779 | nucleotidyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0002020 | protease binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:190273 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:104462 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005760 | gamma DNA polymerase complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005760 | gamma DNA polymerase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | HDA | J:100953 | |||||||||
| Cellular Component | GO:0005759 | mitochondrial matrix | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042645 | mitochondrial nucleoid | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:218220 | |||||||||
| Cellular Component | GO:0043195 | terminal bouton | ISO | J:155856 | |||||||||
| Biological Process | GO:0008340 | determination of adult lifespan | IMP | J:91077 | |||||||||
| Biological Process | GO:0006260 | DNA replication | IEA | J:60000 | |||||||||
| Biological Process | GO:0006260 | DNA replication | IEA | J:72247 | |||||||||
| Biological Process | GO:0006261 | DNA-templated DNA replication | ISO | J:164563 | |||||||||
| Biological Process | GO:0006264 | mitochondrial DNA replication | ISO | J:164563 | |||||||||
| Biological Process | GO:0006264 | mitochondrial DNA replication | IBA | J:265628 | |||||||||
| Biological Process | GO:0006264 | mitochondrial DNA replication | IMP | J:99794 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||