|
Symbol Name ID |
Lims1
LIM and senescent cell antigen-like domains 1 MGI:1195263 |
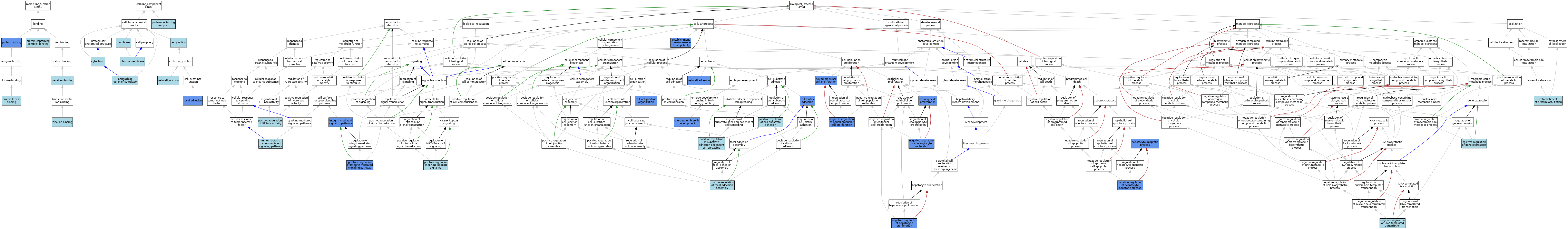
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:80923 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94254 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:80923 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:82868 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:80923 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Biological Process | GO:0098609 | cell-cell adhesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0098609 | cell-cell adhesion | IMP | J:99800 | |||||||||
| Biological Process | GO:0098609 | cell-cell adhesion | IMP | J:99800 | |||||||||
| Biological Process | GO:0045216 | cell-cell junction organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0045216 | cell-cell junction organization | IGI | J:167114 | |||||||||
| Biological Process | GO:0007160 | cell-matrix adhesion | IMP | J:99800 | |||||||||
| Biological Process | GO:0007160 | cell-matrix adhesion | IMP | J:99800 | |||||||||
| Biological Process | GO:1990705 | cholangiocyte proliferation | IGI | J:207543 | |||||||||
| Biological Process | GO:0043009 | chordate embryonic development | IMP | J:97635 | |||||||||
| Biological Process | GO:0045184 | establishment of protein localization | ISO | J:164563 | |||||||||
| Biological Process | GO:0007163 | establishment or maintenance of cell polarity | IMP | J:99800 | |||||||||
| Biological Process | GO:0007163 | establishment or maintenance of cell polarity | IMP | J:99800 | |||||||||
| Biological Process | GO:0097284 | hepatocyte apoptotic process | IGI | J:207543 | |||||||||
| Biological Process | GO:0007229 | integrin-mediated signaling pathway | IGI | J:167114 | |||||||||
| Biological Process | GO:1904055 | negative regulation of cholangiocyte proliferation | IGI | J:207543 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:1903944 | negative regulation of hepatocyte apoptotic process | IGI | J:207543 | |||||||||
| Biological Process | GO:2000346 | negative regulation of hepatocyte proliferation | IGI | J:207543 | |||||||||
| Biological Process | GO:2000178 | negative regulation of neural precursor cell proliferation | IGI | J:208112 | |||||||||
| Biological Process | GO:2000178 | negative regulation of neural precursor cell proliferation | IGI | J:208112 | |||||||||
| Biological Process | GO:0061351 | neural precursor cell proliferation | IGI | J:208112 | |||||||||
| Biological Process | GO:0061351 | neural precursor cell proliferation | IGI | J:208112 | |||||||||
| Biological Process | GO:0010811 | positive regulation of cell-substrate adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0051894 | positive regulation of focal adhesion assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:2001046 | positive regulation of integrin-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:2001046 | positive regulation of integrin-mediated signaling pathway | IGI | J:167114 | |||||||||
| Biological Process | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | ISO | J:164563 | |||||||||
| Biological Process | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | IBA | J:265628 | |||||||||
| Biological Process | GO:0033209 | tumor necrosis factor-mediated signaling pathway | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||