|
Symbol Name ID |
Tln1
talin 1 MGI:1099832 |
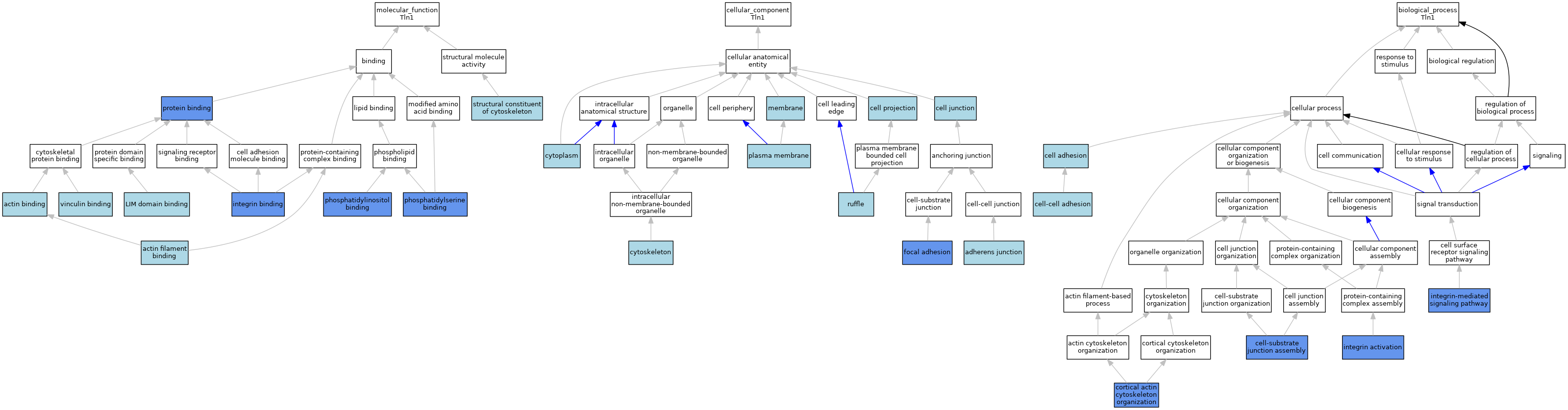
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0051015 | actin filament binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | IMP | J:158602 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005178 | integrin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0030274 | LIM domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035091 | phosphatidylinositol binding | IMP | J:158602 | |||||||||
| Molecular Function | GO:0001786 | phosphatidylserine binding | IMP | J:158602 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245286 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:116247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245251 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:160366 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:247412 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200373 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245362 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:180707 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245320 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:118784 | |||||||||
| Molecular Function | GO:0005200 | structural constituent of cytoskeleton | IEA | J:72247 | |||||||||
| Molecular Function | GO:0017166 | vinculin binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:80557 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0001726 | ruffle | IEA | J:72247 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IEA | J:72247 | |||||||||
| Biological Process | GO:0098609 | cell-cell adhesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0007044 | cell-substrate junction assembly | IMP | J:84472 | |||||||||
| Biological Process | GO:0030866 | cortical actin cytoskeleton organization | IMP | J:84472 | |||||||||
| Biological Process | GO:0033622 | integrin activation | IMP | J:158602 | |||||||||
| Biological Process | GO:0007229 | integrin-mediated signaling pathway | IMP | J:158602 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||