|
Symbol Name ID |
Mtm1
X-linked myotubular myopathy gene 1 MGI:1099452 |
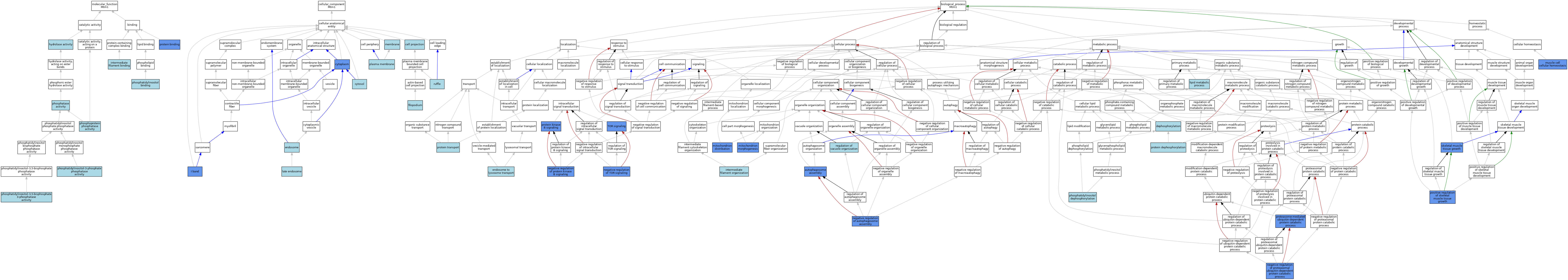
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0019215 | intermediate filament binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016791 | phosphatase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0035091 | phosphatidylinositol binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004438 | phosphatidylinositol-3-phosphate phosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004721 | phosphoprotein phosphatase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:171846 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:203880 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:175210 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030175 | filopodium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031674 | I band | IDA | J:136876 | |||||||||
| Cellular Component | GO:0005770 | late endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0001726 | ruffle | ISO | J:164563 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IMP | J:201107 | |||||||||
| Biological Process | GO:0016311 | dephosphorylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0008333 | endosome to lysosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0045109 | intermediate filament organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0048311 | mitochondrion distribution | ISO | J:164563 | |||||||||
| Biological Process | GO:0048311 | mitochondrion distribution | IMP | J:171846 | |||||||||
| Biological Process | GO:0048311 | mitochondrion distribution | IBA | J:265628 | |||||||||
| Biological Process | GO:0048311 | mitochondrion distribution | IMP | J:136876 | |||||||||
| Biological Process | GO:0070584 | mitochondrion morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0070584 | mitochondrion morphogenesis | IMP | J:171846 | |||||||||
| Biological Process | GO:0070584 | mitochondrion morphogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0046716 | muscle cell cellular homeostasis | IMP | J:194971 | |||||||||
| Biological Process | GO:0046716 | muscle cell cellular homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:0046716 | muscle cell cellular homeostasis | IMP | J:81791 | |||||||||
| Biological Process | GO:1902902 | negative regulation of autophagosome assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:1902902 | negative regulation of autophagosome assembly | IMP | J:201107 | |||||||||
| Biological Process | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | IMP | J:201107 | |||||||||
| Biological Process | GO:0051898 | negative regulation of protein kinase B signaling | IMP | J:201107 | |||||||||
| Biological Process | GO:0032007 | negative regulation of TOR signaling | IMP | J:201107 | |||||||||
| Biological Process | GO:0046856 | phosphatidylinositol dephosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0048633 | positive regulation of skeletal muscle tissue growth | IMP | J:201107 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | IMP | J:201107 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043491 | protein kinase B signaling | IMP | J:201107 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0044088 | regulation of vacuole organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0048630 | skeletal muscle tissue growth | IMP | J:201107 | |||||||||
| Biological Process | GO:0031929 | TOR signaling | IMP | J:201107 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||