|
Symbol Name ID |
Wasf2
WASP family, member 2 MGI:1098641 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
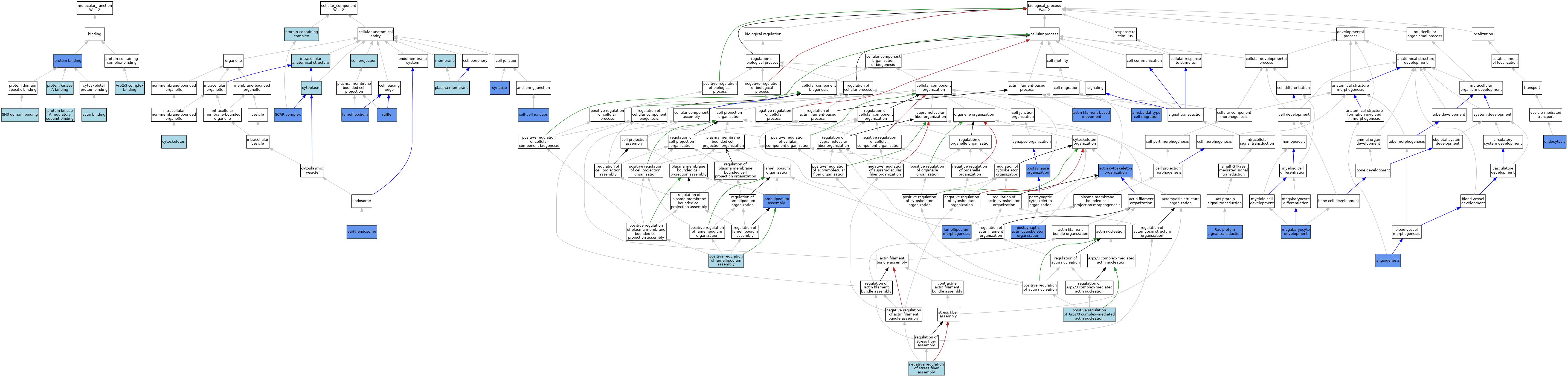
| Molecular Function | GO:0003779 | actin binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003779 | actin binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0071933 | Arp2/3 complex binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:164892 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:130482 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114108 | |||||||||
| Molecular Function | GO:0051018 | protein kinase A binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0034237 | protein kinase A regulatory subunit binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | IDA | J:205441 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005769 | early endosome | IDA | J:81687 | |||||||||
| Cellular Component | GO:0005622 | intracellular anatomical structure | ISO | J:117719 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IDA | J:149119 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | ISO | J:117719 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001726 | ruffle | IDA | J:81687 | |||||||||
| Cellular Component | GO:0031209 | SCAR complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031209 | SCAR complex | IDA | J:302461 | |||||||||
| Cellular Component | GO:0031209 | SCAR complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0045202 | synapse | EXP | J:263402 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:263402 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | IMP | J:84642 | |||||||||
| Biological Process | GO:0030048 | actin filament-based movement | IMP | J:84642 | |||||||||
| Biological Process | GO:0001667 | ameboidal-type cell migration | IMP | J:84651 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IMP | J:84651 | |||||||||
| Biological Process | GO:0006897 | endocytosis | IDA | J:81687 | |||||||||
| Biological Process | GO:0030032 | lamellipodium assembly | IMP | J:84651 | |||||||||
| Biological Process | GO:0072673 | lamellipodium morphogenesis | IMP | J:149119 | |||||||||
| Biological Process | GO:0035855 | megakaryocyte development | IMP | J:149119 | |||||||||
| Biological Process | GO:0051497 | negative regulation of stress fiber assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | IBA | J:265628 | |||||||||
| Biological Process | GO:0010592 | positive regulation of lamellipodium assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0099173 | postsynapse organization | IMP | J:263380 | |||||||||
| Biological Process | GO:0099173 | postsynapse organization | IDA | J:263380 | |||||||||
| Biological Process | GO:0099173 | postsynapse organization | IDA | J:263380 | |||||||||
| Biological Process | GO:0098974 | postsynaptic actin cytoskeleton organization | IMP | J:263380 | |||||||||
| Biological Process | GO:0098974 | postsynaptic actin cytoskeleton organization | IDA | J:263380 | |||||||||
| Biological Process | GO:0098974 | postsynaptic actin cytoskeleton organization | IDA | J:263380 | |||||||||
| Biological Process | GO:0016601 | Rac protein signal transduction | IMP | J:84642 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||