|
Symbol Name ID |
Kif5c
kinesin family member 5C MGI:1098269 |
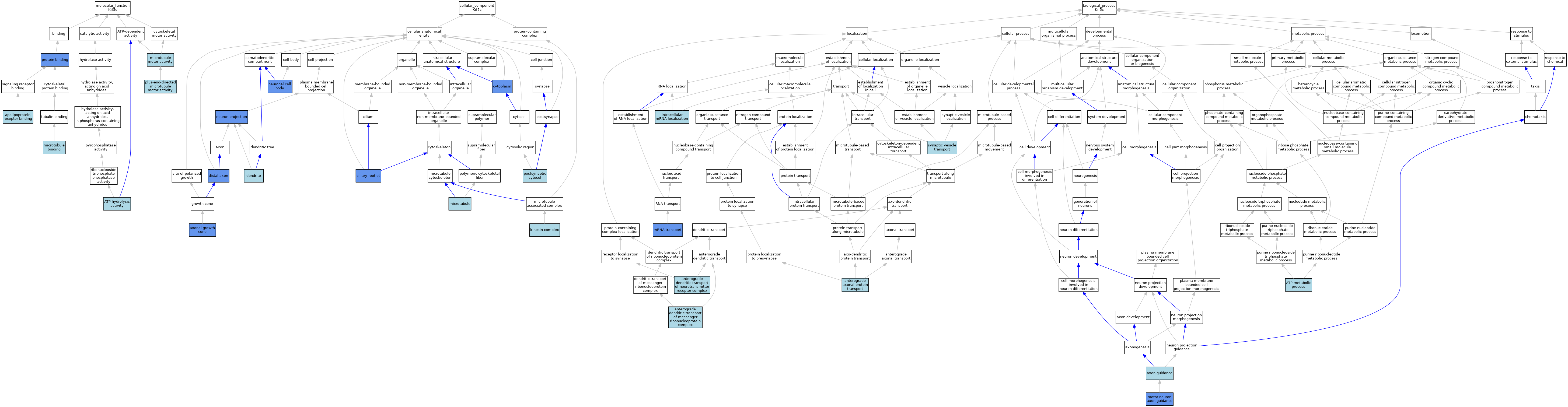
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0034190 | apolipoprotein receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003777 | microtubule motor activity | TAS | J:64206 | |||||||||
| Molecular Function | GO:0008574 | plus-end-directed microtubule motor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:235257 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:176747 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:59015 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:64206 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:64206 | |||||||||
| Cellular Component | GO:0044295 | axonal growth cone | IDA | J:259119 | |||||||||
| Cellular Component | GO:0035253 | ciliary rootlet | IDA | J:102734 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:102734 | |||||||||
| Cellular Component | GO:0030425 | dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0150034 | distal axon | IDA | J:259119 | |||||||||
| Cellular Component | GO:0005871 | kinesin complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005871 | kinesin complex | ISS | J:64206 | |||||||||
| Cellular Component | GO:0005871 | kinesin complex | TAS | J:64206 | |||||||||
| Cellular Component | GO:0005874 | microtubule | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | IDA | J:104505 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:259119 | |||||||||
| Cellular Component | GO:0099524 | postsynaptic cytosol | ISO | J:155856 | |||||||||
| Biological Process | GO:0099641 | anterograde axonal protein transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0098964 | anterograde dendritic transport of messenger ribonucleoprotein complex | ISO | J:155856 | |||||||||
| Biological Process | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0046034 | ATP metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0007411 | axon guidance | IBA | J:265628 | |||||||||
| Biological Process | GO:0008298 | intracellular mRNA localization | ISO | J:155856 | |||||||||
| Biological Process | GO:0008045 | motor neuron axon guidance | IMP | J:64206 | |||||||||
| Biological Process | GO:0051028 | mRNA transport | IMP | J:155254 | |||||||||
| Biological Process | GO:0048489 | synaptic vesicle transport | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||