|
Symbol Name ID |
Kif7
kinesin family member 7 MGI:1098239 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
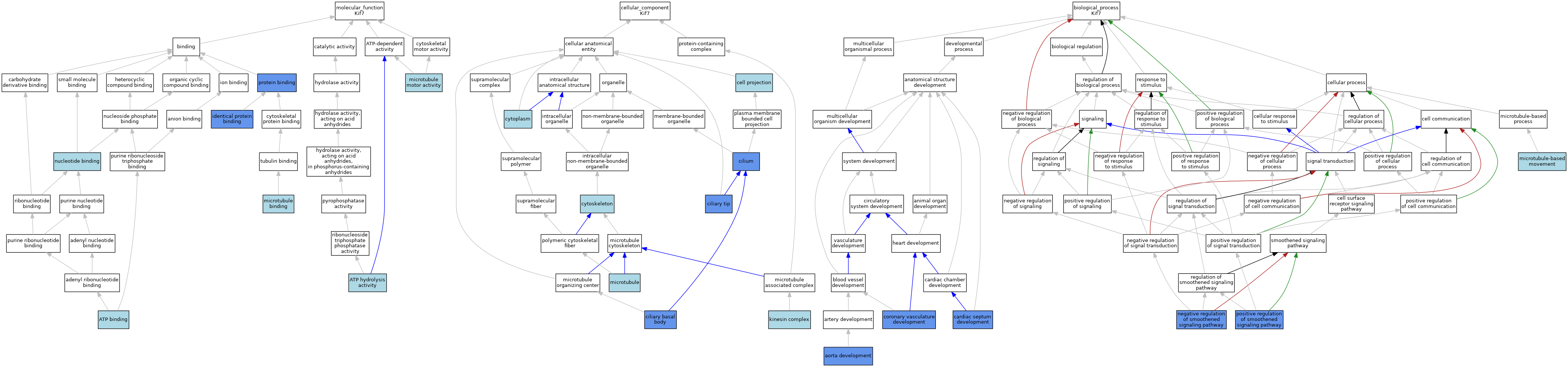
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:219388 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003777 | microtubule motor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003777 | microtubule motor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:218232 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:152259 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:232111 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0036064 | ciliary basal body | IDA | J:218232 | |||||||||
| Cellular Component | GO:0097542 | ciliary tip | IDA | J:255486 | |||||||||
| Cellular Component | GO:0005929 | cilium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005929 | cilium | IDA | J:151965 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005871 | kinesin complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005874 | microtubule | IBA | J:265628 | |||||||||
| Biological Process | GO:0035904 | aorta development | IMP | J:222159 | |||||||||
| Biological Process | GO:0003279 | cardiac septum development | IMP | J:222159 | |||||||||
| Biological Process | GO:0060976 | coronary vasculature development | IMP | J:222159 | |||||||||
| Biological Process | GO:0007018 | microtubule-based movement | IBA | J:265628 | |||||||||
| Biological Process | GO:0045879 | negative regulation of smoothened signaling pathway | IMP | J:152259 | |||||||||
| Biological Process | GO:0045879 | negative regulation of smoothened signaling pathway | IMP | J:152551 | |||||||||
| Biological Process | GO:0045879 | negative regulation of smoothened signaling pathway | IMP | J:151965 | |||||||||
| Biological Process | GO:0045880 | positive regulation of smoothened signaling pathway | IMP | J:152259 | |||||||||
| Biological Process | GO:0045880 | positive regulation of smoothened signaling pathway | IMP | J:151965 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||