|
Symbol Name ID |
Casp7
caspase 7 MGI:109383 |
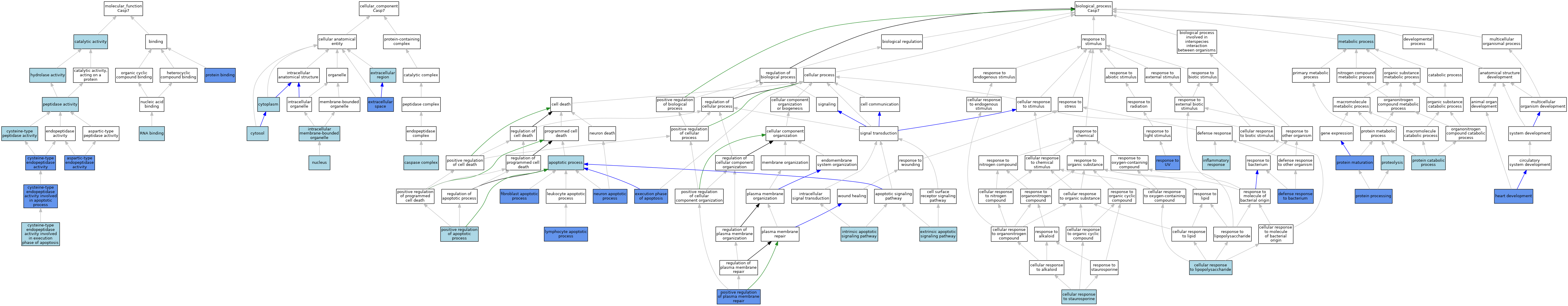
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004190 | aspartic-type endopeptidase activity | IDA | J:326897 | |||||||||
| Molecular Function | GO:0004190 | aspartic-type endopeptidase activity | IDA | J:93470 | |||||||||
| Molecular Function | GO:0003824 | catalytic activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004197 | cysteine-type endopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004197 | cysteine-type endopeptidase activity | IDA | J:180563 | |||||||||
| Molecular Function | GO:0004197 | cysteine-type endopeptidase activity | IDA | J:326897 | |||||||||
| Molecular Function | GO:0004197 | cysteine-type endopeptidase activity | IDA | J:326897 | |||||||||
| Molecular Function | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process | IDA | J:306586 | |||||||||
| Molecular Function | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process | ISO | J:164563 | |||||||||
| Molecular Function | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process | IDA | J:101402 | |||||||||
| Molecular Function | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis | ISO | J:164563 | |||||||||
| Molecular Function | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008234 | cysteine-type peptidase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | RCA | J:84168 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:180563 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0008303 | caspase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:326897 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:326897 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0071222 | cellular response to lipopolysaccharide | ISO | J:164563 | |||||||||
| Biological Process | GO:0072734 | cellular response to staurosporine | ISO | J:164563 | |||||||||
| Biological Process | GO:0042742 | defense response to bacterium | IDA | J:326897 | |||||||||
| Biological Process | GO:0097194 | execution phase of apoptosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0097194 | execution phase of apoptosis | IBA | J:265628 | |||||||||
| Biological Process | GO:0097194 | execution phase of apoptosis | IGI | J:105496 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | NAS | J:320305 | |||||||||
| Biological Process | GO:0044346 | fibroblast apoptotic process | IGI | J:105496 | |||||||||
| Biological Process | GO:0007507 | heart development | IGI | J:105496 | |||||||||
| Biological Process | GO:0007507 | heart development | IGI | J:105496 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | NAS | J:320305 | |||||||||
| Biological Process | GO:0097193 | intrinsic apoptotic signaling pathway | NAS | J:320305 | |||||||||
| Biological Process | GO:0070227 | lymphocyte apoptotic process | IMP | J:146876 | |||||||||
| Biological Process | GO:0008152 | metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0051402 | neuron apoptotic process | IDA | J:93470 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:1905686 | positive regulation of plasma membrane repair | IDA | J:326897 | |||||||||
| Biological Process | GO:0030163 | protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0030163 | protein catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0051604 | protein maturation | ISO | J:164563 | |||||||||
| Biological Process | GO:0051604 | protein maturation | IDA | J:326897 | |||||||||
| Biological Process | GO:0051604 | protein maturation | IDA | J:326897 | |||||||||
| Biological Process | GO:0016485 | protein processing | IDA | J:93470 | |||||||||
| Biological Process | GO:0006508 | proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006508 | proteolysis | RCA | J:84168 | |||||||||
| Biological Process | GO:0009411 | response to UV | IGI | J:105496 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||